Advertisement
ArticleEndocrinology Free access | 10.1172/JCI15276
Defective insulin secretion in pancreatic β cells lacking type 1 IGF receptor
Shouhong Xuan,1 Tadahiro Kitamura,2 Jun Nakae,2 Katerina Politi,1 Yoshiaki Kido,2 Peter E. Fisher,3 Manrico Morroni,4 Saverio Cinti,4 Morris F. White,5 Pedro L. Herrera,6 Domenico Accili,2 and Argiris Efstratiadis1
1Department of Genetics and Development,2 Department of Medicine, and3 Department of Pathology, College of Physicians & Surgeons, Columbia University, New York, New York, USA4 Department of Anatomy, University of Ancona Medical School, Ancona, Italy5 Howard Hughes Medical Institute, and Joslin Diabetes Center, Harvard University, Boston, Massachusetts, USA6 Department of Morphology, University of Geneva, Geneva, Switzerland
Address correspondence to: Domenico Accili, Berrie Research Pavilion, 1150 Saint Nicholas Avenue Room 238, New York, New York 10032, USA. Phone: (212) 851-5332; Fax (212) 851-5331; E-mail: da230@columbia.edu.
Find articles by Xuan, S. in: PubMed | Google Scholar
1Department of Genetics and Development,2 Department of Medicine, and3 Department of Pathology, College of Physicians & Surgeons, Columbia University, New York, New York, USA4 Department of Anatomy, University of Ancona Medical School, Ancona, Italy5 Howard Hughes Medical Institute, and Joslin Diabetes Center, Harvard University, Boston, Massachusetts, USA6 Department of Morphology, University of Geneva, Geneva, Switzerland
Address correspondence to: Domenico Accili, Berrie Research Pavilion, 1150 Saint Nicholas Avenue Room 238, New York, New York 10032, USA. Phone: (212) 851-5332; Fax (212) 851-5331; E-mail: da230@columbia.edu.
Find articles by Kitamura, T. in: PubMed | Google Scholar
1Department of Genetics and Development,2 Department of Medicine, and3 Department of Pathology, College of Physicians & Surgeons, Columbia University, New York, New York, USA4 Department of Anatomy, University of Ancona Medical School, Ancona, Italy5 Howard Hughes Medical Institute, and Joslin Diabetes Center, Harvard University, Boston, Massachusetts, USA6 Department of Morphology, University of Geneva, Geneva, Switzerland
Address correspondence to: Domenico Accili, Berrie Research Pavilion, 1150 Saint Nicholas Avenue Room 238, New York, New York 10032, USA. Phone: (212) 851-5332; Fax (212) 851-5331; E-mail: da230@columbia.edu.
Find articles by Nakae, J. in: PubMed | Google Scholar
1Department of Genetics and Development,2 Department of Medicine, and3 Department of Pathology, College of Physicians & Surgeons, Columbia University, New York, New York, USA4 Department of Anatomy, University of Ancona Medical School, Ancona, Italy5 Howard Hughes Medical Institute, and Joslin Diabetes Center, Harvard University, Boston, Massachusetts, USA6 Department of Morphology, University of Geneva, Geneva, Switzerland
Address correspondence to: Domenico Accili, Berrie Research Pavilion, 1150 Saint Nicholas Avenue Room 238, New York, New York 10032, USA. Phone: (212) 851-5332; Fax (212) 851-5331; E-mail: da230@columbia.edu.
Find articles by Politi, K. in: PubMed | Google Scholar
1Department of Genetics and Development,2 Department of Medicine, and3 Department of Pathology, College of Physicians & Surgeons, Columbia University, New York, New York, USA4 Department of Anatomy, University of Ancona Medical School, Ancona, Italy5 Howard Hughes Medical Institute, and Joslin Diabetes Center, Harvard University, Boston, Massachusetts, USA6 Department of Morphology, University of Geneva, Geneva, Switzerland
Address correspondence to: Domenico Accili, Berrie Research Pavilion, 1150 Saint Nicholas Avenue Room 238, New York, New York 10032, USA. Phone: (212) 851-5332; Fax (212) 851-5331; E-mail: da230@columbia.edu.
Find articles by Kido, Y. in: PubMed | Google Scholar
1Department of Genetics and Development,2 Department of Medicine, and3 Department of Pathology, College of Physicians & Surgeons, Columbia University, New York, New York, USA4 Department of Anatomy, University of Ancona Medical School, Ancona, Italy5 Howard Hughes Medical Institute, and Joslin Diabetes Center, Harvard University, Boston, Massachusetts, USA6 Department of Morphology, University of Geneva, Geneva, Switzerland
Address correspondence to: Domenico Accili, Berrie Research Pavilion, 1150 Saint Nicholas Avenue Room 238, New York, New York 10032, USA. Phone: (212) 851-5332; Fax (212) 851-5331; E-mail: da230@columbia.edu.
Find articles by Fisher, P. in: PubMed | Google Scholar
1Department of Genetics and Development,2 Department of Medicine, and3 Department of Pathology, College of Physicians & Surgeons, Columbia University, New York, New York, USA4 Department of Anatomy, University of Ancona Medical School, Ancona, Italy5 Howard Hughes Medical Institute, and Joslin Diabetes Center, Harvard University, Boston, Massachusetts, USA6 Department of Morphology, University of Geneva, Geneva, Switzerland
Address correspondence to: Domenico Accili, Berrie Research Pavilion, 1150 Saint Nicholas Avenue Room 238, New York, New York 10032, USA. Phone: (212) 851-5332; Fax (212) 851-5331; E-mail: da230@columbia.edu.
Find articles by Morroni, M. in: PubMed | Google Scholar
1Department of Genetics and Development,2 Department of Medicine, and3 Department of Pathology, College of Physicians & Surgeons, Columbia University, New York, New York, USA4 Department of Anatomy, University of Ancona Medical School, Ancona, Italy5 Howard Hughes Medical Institute, and Joslin Diabetes Center, Harvard University, Boston, Massachusetts, USA6 Department of Morphology, University of Geneva, Geneva, Switzerland
Address correspondence to: Domenico Accili, Berrie Research Pavilion, 1150 Saint Nicholas Avenue Room 238, New York, New York 10032, USA. Phone: (212) 851-5332; Fax (212) 851-5331; E-mail: da230@columbia.edu.
Find articles by Cinti, S. in: PubMed | Google Scholar
1Department of Genetics and Development,2 Department of Medicine, and3 Department of Pathology, College of Physicians & Surgeons, Columbia University, New York, New York, USA4 Department of Anatomy, University of Ancona Medical School, Ancona, Italy5 Howard Hughes Medical Institute, and Joslin Diabetes Center, Harvard University, Boston, Massachusetts, USA6 Department of Morphology, University of Geneva, Geneva, Switzerland
Address correspondence to: Domenico Accili, Berrie Research Pavilion, 1150 Saint Nicholas Avenue Room 238, New York, New York 10032, USA. Phone: (212) 851-5332; Fax (212) 851-5331; E-mail: da230@columbia.edu.
Find articles by White, M. in: PubMed | Google Scholar
1Department of Genetics and Development,2 Department of Medicine, and3 Department of Pathology, College of Physicians & Surgeons, Columbia University, New York, New York, USA4 Department of Anatomy, University of Ancona Medical School, Ancona, Italy5 Howard Hughes Medical Institute, and Joslin Diabetes Center, Harvard University, Boston, Massachusetts, USA6 Department of Morphology, University of Geneva, Geneva, Switzerland
Address correspondence to: Domenico Accili, Berrie Research Pavilion, 1150 Saint Nicholas Avenue Room 238, New York, New York 10032, USA. Phone: (212) 851-5332; Fax (212) 851-5331; E-mail: da230@columbia.edu.
Find articles by Herrera, P. in: PubMed | Google Scholar
1Department of Genetics and Development,2 Department of Medicine, and3 Department of Pathology, College of Physicians & Surgeons, Columbia University, New York, New York, USA4 Department of Anatomy, University of Ancona Medical School, Ancona, Italy5 Howard Hughes Medical Institute, and Joslin Diabetes Center, Harvard University, Boston, Massachusetts, USA6 Department of Morphology, University of Geneva, Geneva, Switzerland
Address correspondence to: Domenico Accili, Berrie Research Pavilion, 1150 Saint Nicholas Avenue Room 238, New York, New York 10032, USA. Phone: (212) 851-5332; Fax (212) 851-5331; E-mail: da230@columbia.edu.
Find articles by Accili, D. in: PubMed | Google Scholar
1Department of Genetics and Development,2 Department of Medicine, and3 Department of Pathology, College of Physicians & Surgeons, Columbia University, New York, New York, USA4 Department of Anatomy, University of Ancona Medical School, Ancona, Italy5 Howard Hughes Medical Institute, and Joslin Diabetes Center, Harvard University, Boston, Massachusetts, USA6 Department of Morphology, University of Geneva, Geneva, Switzerland
Address correspondence to: Domenico Accili, Berrie Research Pavilion, 1150 Saint Nicholas Avenue Room 238, New York, New York 10032, USA. Phone: (212) 851-5332; Fax (212) 851-5331; E-mail: da230@columbia.edu.
Find articles by Efstratiadis, A. in: PubMed | Google Scholar
Published October 1, 2002 - More info
J Clin Invest. 2002;110(7):1011–1019. https://doi.org/10.1172/JCI15276.
© 2002 The American Society for Clinical Investigation
Received: February 2, 2002; Accepted: August 9, 2002
-
Abstract
Defective insulin secretion is a feature of type 2 diabetes that results from inadequate compensatory increase of β cell mass and impaired glucose-dependent insulin release. β cell proliferation and secretion are thought to be regulated by signaling through receptor tyrosine kinases. In this regard, we sought to examine the potential proliferative and/or antiapoptotic role of IGFs in β cells by tissue-specific conditional mutagenesis ablating type 1 IGF receptor (IGF1R) signaling. Unexpectedly, lack of functional IGF1R did not affect β cell mass, but resulted in age-dependent impairment of glucose tolerance, associated with a decrease of glucose- and arginine-dependent insulin release. These observations reveal a requirement of IGF1R-mediated signaling for insulin secretion.
-
Introduction
The ability of pancreatic β cells to synthesize, store, and release insulin in response to variations in circulating metabolite levels and intracellular glucose metabolism is regulated by changes in ATP/ADP ratios resulting in Ca2+ mobilization (1). Alterations of this sensing loop occur early in the pathogenesis of type 2 diabetes, but are initially compensated by an increase of β cell mass (2). In this respect, pancreatic β cells appear to differ from other terminally differentiated cell types by retaining their ability to proliferate, as demonstrated in both physiological conditions (growth, gestation) and disease states (obesity, insulin resistance) (3). In addition to presumptive proliferation of existing β cells, there is evidence for β cell neogenesis from undifferentiated progenitors, apparently arising from the epithelial lining of pancreatic ducts (4–8).
The factors inducing β cell proliferation under normal or pathological conditions are largely unknown, although some evidence exists about the involvement of FGFs (9), HGFs (10), and placental lactogen (11). Moreover, signaling by receptor tyrosine kinases has been implicated as a regulatory mechanism in both β cell proliferation (12–14) and insulin release (9, 15–19). In particular, insulin/IGF signaling through insulin receptor substrate (IRS) and phosphoinositide 3-kinase (PI 3-kinase) appears to regulate several aspects of β cell function. Thus, ablation of the insulin/IGF receptor substrate IRS-2 impairs β cell proliferation (20, 21), whereas ablation of p70s6k1, an Akt substrate, is associated with a decrease in β cell size (22). Conversely, overexpression of a constitutively active mutant Akt1 increases β cell mass and protects from streptozotocin-induced diabetes (23, 24). However, mutations affecting the components of the IRS-2→Akt→p70s6k1 axis do not perturb glucose sensing or insulin production. In contrast, mutations of Insulin receptor (Ir) (25) or Irs1 (26) impair insulin synthesis and secretion mediated by PI 3-kinase–dependent pathways (17, 19, 26). These observations suggest that signals regulating β cell proliferation and insulin secretion diverge downstream of PI 3-kinase; i.e., Akt is not the sole effector of PI 3-kinase. Nevertheless, the role of growth factor signaling through PI 3-kinase as related to insulin secretion remains poorly understood. Additional difficulties arise because, against expectation, knockout of the gene encoding the p85α subunit of PI 3-kinase results in increased insulin secretion (27), consistent with studies using PI 3-kinase inhibitors (28). Because of the known relationship between IGF and PI 3-kinase signaling and results indicating that IGFs inhibit insulin secretion (29–31), we sought to examine in mutant mice the contribution of type 1 IGF receptor (IGF1R) function to insulin secretion and β cell proliferation, which could differ from that of the IR pathway.
Although the roles of IGFs in β cells have not been completely elucidated, these ligands and their cognate receptors are both expressed in this tissue (32–35). Interestingly, the developmental wave of β cell apoptosis observed in neonatal rats (36) is associated with a decrease in IGF-2 expression (32, 37), leading to the suggestion that the IGFs have an antiapoptotic function in β cells. On the other hand, overexpression of IGF-2 in β cells (38), but not in liver (39, 40), results in diabetes, presumably through increased insulin production with concomitant downregulation of insulin sensitivity.
A potential role for IGF1R acting as a mediator of β cell proliferation through IRS-2 has been implied by observations indicating that the β cell failure observed in Irs2–/– mice is exacerbated by Igf1r haploinsufficiency (13). However, generalized lack of IGF1R is lethal in the immediate postnatal period (41), thus precluding an extensive analysis of the role of IGF signaling in β cell physiology. To circumvent this limitation and analyze the role of IGF1R in β cells, we have used conditional mutagenesis with the cre/loxP site-specific recombination system to generate mice lacking IGF1R specifically in pancreatic β cells. Here we show that these mutant mice are glucose intolerant and exhibit an insulin secretion defect.
-
Methods
Mice. Animals carrying a floxed Igf1r allele (Igf1rlox) (42) or a null Igf1r allele (Igf1r+/–) (41) and transgenic mice expressing the Cre recombinase under the transcriptional control of the rat insulin-2 promoter (InsPr-Cre) (43) have been described previously. A mating program with Igf1r+/–, Igf1rlox/+, and InsPr-Cre mice was used to generate progeny of five genotypes: Igf1r+/–, Igf1rlox/+, Igf1rlox/–, Igf1rΔlox/–, and Igf1rΔlox/+ (in which exon 3 sequences have been deleted as a result of Cre-mediated recombination). PCR analysis was used for genotyping. The wild-type, null, and Igf1rlox alleles were detected with primer 5′ CTTCCCAGCTTGCTACTCTAGG 3′ (P1, forward, located upstream of the second loxP site) and 5′ CAGGCTTGCAATGAGACATGGG 3′ (P2, reverse, located downstream of the same loxP site) (Figure 1). The Igf1rΔlox allele was detected using a different forward primer, 5′ TGAGACGTAGCGAGATTGCTGTA 3′, located 5′ of the loxP site in intron 2 (P3 in Figure 1), and the reverse primer, P2. The products of the three alleles were 120 bp (wild-type or null allele), 220 bp (Igf1rlox), and 320 bp (Igf1rΔlox). PCR amplification conditions were 30 cycles at 94°C for 45 seconds, 61°C for 45 seconds, and 72°C for 1 minute.
 Figure 1
Figure 1Generation of a conditional IGF1R knockout in β cells. (a) A diagram of the Igf1r locus around exon 3 (filled square) is shown, followed by diagrams of the null (Igf1r+/–) (41), floxed (Igf1rlox) (42), and recombined floxed (Igf1rΔlox) (42) alleles. The positions of the P1, P2, and P3 primers (arrows) used for genotyping are indicated. (b) PCR analyses using DNA from islets and control tissues. The lanes labeled – and + correspond to the negative and positive controls. Primers P1 and P2 yield a product of approximately 120 bp in WT and null Igf1r alleles, and 220 bp in the Igf1rlox allele. In the Igf1rΔlox allele, excision of exon 3 enables amplification to occur between primers P3 and P2, yielding 320 bp. In the lanes to the right of the markers, DNA from different tissues of Igf1rΔlox/– mice was used. Br, brain; Li, liver; and Ki, kidney. (c) Western blot analysis of IGF1R expression. We obtained protein extracts from wild-type mouse embryonic fibroblasts or from fibroblasts derived from Igf1r–/– mice as positive and negative controls (first two lanes on the left). We purified islets from wild-type (third lane from the left) and Igf1rΔlox/– mice (right lane). Following detection of IGF1R, the blot was stripped and reprobed with anti-tubulin antiserum. The position of the various bands is indicated on the left of the autoradiogram.
Ab’s and Western blot analysis. An antipeptide antiserum against the carboxy-terminal domain of the IGF1R protein (C20; Santa Cruz Biotechnology Inc., Santa Cruz, California, USA) was used for Western blot analysis as described previously (44). Anti-tubulin antiserum was from Covance (Richmond, Virginia, USA). We employed a Triton extraction buffer for protein isolation and standard methods for Western blot analysis. The intensity of the bands on the autoradiogram was quantified using a Kodak Image Station C400 image analyzer for chemiluminescence.
Phenotypic analysis. Only male mice were used in the analysis, since they are more prone to insulin resistance. Blood glucose levels were determined using an Accucheck glucometer (Boehringer Mannheim GmbH, Mannheim, Germany). Serum insulin was measured by a radioimmunoassay using a rat insulin standard (Linco Research Inc., St. Charles, Missouri, USA). All assays were carried out in duplicate (45).
Intraperitoneal glucose tolerance test. Mice were fasted for 16 hours and then anesthetized. Blood was drawn immediately before and 30, 60, 90, and 120 minutes after intraperitoneal injection of glucose (2 g/kg body weight), and then glucose and insulin levels were determined (45).
Insulin secretion assays. Islets were purified from 6-month-old Igf1rlox/+, Igf1rlox/–, and Igf1rΔlox/– mice by collagenase digestion followed by centrifugation over a Histopaque gradient as described previously (46). For insulin-release experiments, islets were manually picked, incubated in Krebs buffer, and stimulated with varying concentrations of glucose or arginine for 1 hour at 37°C. At the end of the incubation, islets were collected by centrifugation, and the supernatant was assayed for insulin content by radioimmunoassay (46). The cell pellet was assayed for insulin and DNA content to normalize insulin secretion by islet mass.
Immunohistochemical and morphometric analyses. Pancreata were removed from embryonic day 18.5 (E18.5), postnatal day 10 (P10), P30, and P120 Igf1rlox/–, Igf1rΔlox/+, and Igf1rΔlox/– mice, weighed, and fixed overnight in 4% paraformaldehyde. Four-micrometer-thick sections were immunostained for β cells using mouse anti-insulin Ab’s or rabbit anti–GLUT-2 polyclonal Ab (Research Diagnostics, Flanders, New Jersey, USA) and for α cells using mouse anti-glucagon Ab’s (Sigma-Aldrich, St. Louis, Missouri, USA). For immunohistochemistry with anti–GLUT-2 antiserum (Research Diagnostics), pancreata were fixed in Z-fix and incubated at a 1:300 dilution. Immunoreactivity was detected with the ABC system (DAKO Corp., Carpinteria, California, USA). For morphometric analysis of β cell mass, two animals of each genotype were analyzed. For each pancreas, three sections spaced at least 40-μm (E18.5 and P10) and 160-μm apart (P30 and P120) were covered systematically by accumulating images from nonoverlapping fields that were captured with a digital camera (Nikon 950) and analyzed using the NIH Image 1.60 software (Bethesda, Maryland, USA), as described (45). Results were expressed as percentage of the total surveyed pancreatic area occupied by α and β cells. β cell mass was calculated by multiplying β cell area by pancreatic weight. β cell size was calculated from the same sections using a reference standard.
Detection and quantitation of β cell replication. Cellular proliferation was evaluated using immunohistochemistry with three markers: the cell cycle antigen Ki67 (NCL-Ki 67p; Castra, Newcastle upon Tyne, United Kingdom), phosphorylated histone H3 (Upstate Biotechnology Inc., Lake Placid, New York, USA), and 5-bromo-2-deoxyuridine (BrdU; Sigma-Aldrich). For these experiments, pancreata were obtained from E18.5, P10, P30, and P120 Igf1rlox/+, Igf1rlox/–, Igf1rΔlox/+, and Igf1rΔlox/– mice. BrdU was injected intraperitoneally (0.1 gm/kg) 2 hours before sacrifice into mothers at day 18.5 of pregnancy or P10, P20, and P30 Igf1rlox/+, Igf1rlox/–, Igf1rΔlox/+, and Igf1rΔlox/– mice. For double-staining with anti-BrdU or anti-Ki67 and anti-insulin Ab’s, guinea pig anti-insulin Ab was used. Quantitation of β cell replication was performed by counting Ki67-positive cells in three to five sections spaced more than 40 μm (E18.5 and P10) or 160 μm apart (P30 and P120) in each pancreas. The mean value of Ki67-positive cells was multiplied by β cell mass as calculated above to obtain an arbitrary Ki67 labeling index. Apoptosis was measured using the ApopTag peroxidase in situ apoptosis detection kit (S7101; Intergen Co., Purchase, New York, USA) in sections derived from P10, P30, and P120 mice.
Electron microscopy. Two wild-type and two mutant pancreata were fixed in a solution containing 2% glutaraldehyde, 2% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4, for 4 hours at 4°C, post-fixed in 1% osmium tetroxide, dehydrated in ethanol, and embedded in an Epon-Araldite mixture (Multilab Supplies, Fetcham, United Kingdom). Thin sections were obtained with a MT-X ultratome (RMC, Tucson, Arizona, USA), stained with lead citrate, and examined with a CM10 transmission electron microscope (Phillips, Eindhoven, The Netherlands). We examined three islets in each of the two wild-type mice and three or four islets in the two mutant mice. We obtained serial photos at ×1,900 magnification for each islet section and reconstructed the whole islet at a final enlargement of ×4,275. The specimens were examined blindly and independently by two electron microscopists, who analyzed a total of 448 cells in wild-type and 638 cells in mutant mice.
mRNA quantitation by real-time PCR. PolyA+-enriched RNA was isolated from purified islets of Igf1rlox/+, Igf1rlox/–, and Igf1rΔlox/– mice using Micro Fast Track (Invitrogen Corp., Carlsbad, California, USA). Real-time PCR using RNA pooled from three animals per specimen was performed using a Roche Light Cycler PCR instrument and Light-Cycler RT-PCR kit (Roche Perkin-Elmer, Foster City, California, USA). Each reaction was carried out in triplicate.
Microarray analysis. RNA was extracted from islets using the RNeasy Total RNA Isolation Kit (QIAGEN Inc., Valencia, California, USA), precipitated, and quantified by agarose gel electrophoresis using an aliquot. Five micrograms of RNA was used to synthesize double-stranded cDNA (Invitrogen Corp. kit) using a T7 (dT)24 primer (Invitrogen, Carlsbad, California, USA) containing a T7 RNA polymerase initiation site. Biotinylated cRNA was synthesized from the cDNA template using the BioArray High Yield RNA Transcript Labeling Kit (ENZO Diagnostics, Armingdale, New York, USA) and purified with the RNeasy Total RNA Isolation Kit (RNA clean-up protocol; QIAGEN Inc.). After fragmentation in 40 mM Tris-acetate, pH 8.1, 100 mM KOAc, and 30 mM MgOAc, 15 μg fragmented cRNA was hybridized to the MGU74Av2 oligonucleotide microarray (Affymetrix, Santa Clara, California, USA) representative of approximately 12,000 genes. The microarrays were scanned (Affymetrix Scanner), and expression values for the genes were determined using Affymetrix Microarray Suite Version 5.0 and subjected to computer analysis (47).
Statistical analysis. Descriptive statistics, t test for paired data, Kruskal-Wallis test, and ANOVA were performed using the Statsview software.
-
Results
Generation of mice lacking IGF1R in β cells. Tissue-specific conditional mutagenesis requires crosses between Cre-producing and Cre-responding strains of mice. For Cre expression in pancreatic β cells of producer mice we used an InsPr-Cre transgenic line, in which Cre transcription is driven by the rat insulin-2 gene promoter (43). These mice express Cre in β cells as early as embryonic day 11.5 (E11.5), and show very limited extrapancreatic expression in the amygdala (P. Herrera, unpublished observations). As responder mice we used previously described compound heterozygotes (42) derived from matings between heterozygotes carrying an Igf1r null allele (Igf1r+/–) and Igf1rlox/+ conditional mutants carrying a modified Igf1r locus in which exon 3 of the gene is flanked by loxP sites (“floxed”). PCR analysis of DNA extracted from various tissues of progeny carrying the InsPr-Cre transgene and possessing an Igf1rlox/+ genotype showed that DNA excision leaving behind a single loxP site had occurred specifically in pancreatic islets (Figure 1a). Additional Southern blot analysis of DNA from several other tissues from which adequate amounts of DNA could be extracted indicated the presence of an exclusively unrecombined allele (data not shown). The extent of recombination detected in islets was approximately 80%, consistent with the ratio of β/non-β (α, δ, PP, vascular, and connective) cells in normal islets (Figure 1b). Western blot analysis of pancreatic islets with anti-IGF1R antiserum confirmed that the levels of proreceptor and mature β subunit were reduced by 85% in Igf1rΔlox/– mice, consistent with ablation of the protein product exclusively in β cells (Figure 1c). Parallel immunohistochemical analysis yielded consistent results (data not shown). Igf1rΔlox/– mice were born at the expected Mendelian frequency and did not exhibit abnormalities.
Phenotypic characterization of mice lacking IGF1R in β cells. To assess the consequences of IGF1R ablation in β cells on glucose metabolism, we measured glucose and insulin levels in neonates and adult mice in the fasting and fed states at various ages. No differences were detected between mice with Igf1r+/–, Igf1rlox/+, Igf1rlox/–, Igf1rΔlox/+, and Igf1rΔlox/– genotypes (Table 1). Glucose tolerance tests were then performed in adult (6-month-old) mice to assess potential metabolic defects not resulting in overt diabetes. In Igf1r+/– and Igf1rΔlox/+ mice, glucose values rose to approximately 250 mg/dl 30 minutes after intraperitoneal glucose administration and returned to nearly normal values by 2 hours. In contrast, in Igf1rΔlox/– mice glucose values rose to approximately 400 mg/dl and did not decrease appreciably thereafter, consistent with impaired glucose disposal. Igf1rlox/– mice had similar 30-minute glucose values as Igf1rΔlox/– mice, but gradually returned to nearly normal values (Figure 2). These data could be interpreted as indicating that the combination of a single null allele and a floxed allele results in haploinsufficiency for Igf1r and causes a mild impairment of glucose tolerance. The impairment of glucose tolerance in Igf1rlox/– and Igf1rΔlox/– mice is apparently not due to insulin resistance, since insulin tolerance tests failed to demonstrate differences between the two genotypes and wild-type mice (data not shown).We next examined the consequences of ablation of IGF1R function on pancreatic morphology. Immunohistochemical analysis of pancreata from Igf1rΔlox/– mice showed that the islet architecture was normal (Figure 3). Thus, β cell mass was similar in all genotypes examined at various time points, as was mean β cell diameter (Table 2). In addition, the pattern of expression of the β cell–restricted transcription factor Pdx1, as assessed by immunohistochemistry, was similar (data not shown). In contrast, real-time PCR analysis of the expression levels of mRNA encoding the β cell glucose transporter GLUT-2 demonstrated a significant decrease in the islets of Igf1rΔlox/– mice (by approximately 40%) when compared with Igf1rlox/+ and Igf1rlox/– mice (Figure 4a). Immunostaining of pancreatic sections with anti–GLUT-2 antiserum showed decreased immunoreactivity in Igf1rΔlox/– mice (Figure 4b).To determine whether the lack of IGF1R affects β cell proliferation, we measured the number of Ki67-positive β cells in Igf1rlox/–, Igf1rΔlox/+, and Igf1rΔlox/– mice at various ages as an index of mitotic activity. The use of Ki67 for this purpose was validated by staining adjacent sections with two additional proliferation markers, phosphohistone H3 and BrdU, and then analyzing the correlation between the values obtained with different methods. As shown in Figure 5, phosphohistone H3– and BrdU-positive cells showed a highly significant positive correlation with Ki67-positive cells (r2 > 0.8, P < 0.0001). Thus, Ki67 was employed for all subsequent analyses. A developmental analysis showed that the number of β cells undergoing replication decreased sharply with age. However, no differences were detected between the various genotypes under study (Figure 6). These data indicate that lack of IGF1R does not impair β cell replication. Similarly, we observed no differences in the levels of apoptotis ascertained at P10, P20, and P30 using TUNEL staining, although the small number of detectable apoptotic cells did not permit accurate quantitation (data not shown).
 Figure 2
Figure 2Intraperitoneal glucose tolerance tests in mice lacking IGF1R in β cells. Tests were performed in overnight-fasted mice as indicated in Methods. The difference between Igf1rΔlox/– and Igf1rΔlox/+ is significant at all time points except 0. The difference between Igf1rΔlox/– and Igf1rlox/– is statistically significant at times 90 and 120. The difference between Igf1rΔlox/+ and Igf1rlox/– is statistically significant at times 30, 60, and 90 (P < 0.05 by ANOVA). Bars represent SEM.
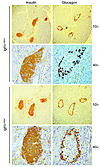 Figure 3
Figure 3Islet morphology in mice lacking IGF1R in β cells. Examples of pancreatic sections from P30 mice of the indicated genotypes that were stained with anti-insulin and anti-glucagon Ab’s are shown.
 Figure 4
Figure 4Analysis of Glut-2 expression. (a) Real-time PCR amplification. mRNA was isolated from Igf1rΔlox/–, Igf1rlox/–, and Igf1rΔlox/+ mice (n = 3 each; equal amounts were pooled). Amplification was performed using a one-step RT-PCR reaction. The amount of RNA in the reaction was normalized using β-actin RNA as a control. *P < 0.05 by ANOVA. (b) Immunohistochemistry with anti-GLUT-2 antiserum. Representative sections of Igf1rΔlox/+ and wild-type mice are shown.
 Figure 5
Figure 5Ki67, histone H3, and BrdU labeling of β cells. Adjacent 4-μm-thick pancreatic section from mice at E18.5, P10, P30, and P120 were costained with anti-insulin Ab’s (to identify β cells) and anti-Ki67, or anti-histone H3 or anti-BrdU Ab’s (to provide an index of proliferation). Positive cells were visually scored, and the number of Ki67-positive cells was plotted as a function of the number of histone H3–positive (upper panel) or BrdU-positive cells (lower panel). Regression analysis was performed using the Statsview software. r2 = 0.9 for the H3/Ki67 and 0.9 for the BrdU/Ki67 correlation (P < 0.0001).
 Figure 6
Figure 6Developmental analysis of Ki67 labeling. A Ki67 labeling index was determined in mice of different ages by multiplying the mean number of Ki67/insulin double-positive cells per section by β cell mass (μg). The values at E18.5 were normalized to 100%. None of the differences observed between the different genotypes are statistically significant. Two mice for each genotype were used at each time point, except for Igf1rlox/+ and Igf1rΔlox/+ at P10, where one mouse was used. Three to five sections per pancreas were scored as described in Methods.
Measurements of insulin secretion in purified islets. To examine whether the conditional mutation under study affects insulin secretion, we purified islets from Igf1rlox/+, Igf1rΔlox/+, and Igf1rΔlox/– mice, which were incubated in the presence of glucose or arginine. As shown in Figure 7a, glucose induced a dose-dependent increase in insulin secretion up to a maximum of 15- to 20-fold over the basal level in islets from Igf1rlox/+ and Igf1rΔlox/+ controls. In Igf1rΔlox/– mutants, glucose-induced insulin secretion was also normal at low glucose concentrations, but decreased by approximately 70% at high concentrations (P < 0.05 by ANOVA). A similar pattern of response was observed in arginine-induced insulin secretion in Igf1rΔlox/– mutant mice (Figure 7b). On the other hand, the islet insulin content was similar in the Igf1rlox/+, Igf1rΔlox/+, and Igf1rΔlox/– genotypes (Figure 7c), indicating that the defective insulin secretion in Igf1rΔlox/– mutants, which apparently leads to impairment of glucose tolerance, is not due to paucity of insulin. To assess whether the observed defect in insulin secretion could be correlated with β cell structure abnormalities, we analyzed β cells from Igf1rΔlox/– mice and wild-type controls by electron microscopy. The results showed that there were no identifiable morphologic abnormalities in β cells lacking IGF1R, except for a hypertrophic Golgi system, indicating that abnormal insulin secretion is not due to overt disruption of cellular architecture and failure to assemble insulin secretory granules (Figure 8).
 Figure 7
Figure 7Insulin secretion and content in purified islets. Insulin release was determined in response to varying glucose (a) and arginine (b) concentrations. For each glucose concentration, ten islets derived from two or three mice of each genotype were used. Three separate experiments were performed and the results were averaged. (c) Insulin content in islets of the indicated genotypes was measured by high-salt buffer extraction followed by radioimmunoassay.
 Figure 8
Figure 8Electron microscopy of β cells. Representative images of β cells in wild-type and mutant mice are shown at ×11,300 magnification.
Analysis of transcripts encoding proteins involved in exocytosis. Considering that the secretion data in isolated islets are consistent with a defect in insulin release, we analyzed the expression of several genes involved in insulin exocytosis using expression profiling with an oligonucleotide microarray. This analysis confirmed that Igf1r mRNA was decreased by approximately 80% in Igf1rΔlox/– mice. However, the assay did not reveal differences in expression levels of several candidate genes, including those encoding the β-granule components synaptotagmin II and III, synaptobrevin 2 (VAMP-2), VAMP-3, and Rab-3a; the cytosolic proteins NSF, 14-3-3, Rab-3 GDI, Mss-4, Munc-18, and annexin II; the plasma membrane proteins SNAP-23, SNAP-25, syntaxin-1A, -3, -4A, -5A, -6; and the synaptic-like vesicles synaptophysin, synapsin I and II. Expression of the sulfonylurea receptor and calmodulin kinase II was also unaffected. Thus, the observed defect in insulin secretion is apparently not due to alterations in members of the insulin exocytotic pathway, at least at the transcriptional level.
-
Discussion
Type 2 diabetes results from combined abnormalities of β cell function and insulin action. Although the exact nature of the β cell defect remains elusive, one of the earliest demonstrable abnormalities in individuals at high risk of developing type 2 diabetes is an exaggerated insulin response to a glucose challenge, suggestive of an intrinsic secretory defect (48, 49). Moreover, unaffected first-degree relatives of type 2 diabetics exhibit abnormalities of the first-phase insulin response to glucose, similar to the defect observed in patients with active disease (50). Notably, β cell secretory defects are exemplified by maturity onset diabetes of the young (51, 52), a genetically heterogeneous, highly penetrant, monogenic form of diabetes caused by mutations in genes that couple glucose metabolism to insulin release (53).
In most type 2 diabetics, defective insulin secretion is compounded with peripheral insulin resistance in various tissues (54). Initially, in response to this resistance, there is a compensatory increase in β cell mass to preserve adequate insulin production. Over time, however, this compensation cannot be sustained. The mechanism of this response and the pathophysiology of its failure in type 2 diabetes remain unclear, but there is some evidence for circulating factors affecting islet proliferation (55). Alternatively, transient oscillations in glucose levels, occurring before the onset of overt hyperglycemia, may be sufficient to promote β cell growth (56), or glucose may have a permissive effect in stimulating β cell proliferation acting in concert with growth factors (12).
The observations that mice lacking IRS-1 develop defective insulin secretion, whereas mice lacking IRS-2 develop impaired β cell proliferation, have raised the question of whether insulin signaling on one hand and IGF signaling on the other serve overlapping or distinct functions related to these phenotypic manifestations (57). In this regard, conditional ablation of Ir in β cells results in a secretory defect similar to that observed in type 2 diabetes (25), but less severe than that observed in IRS-1–deficient β cells (26). Insulin acting through IR appears to be required for regulation of its own synthesis and secretion (17), while the IGFs have been proposed to be involved in the promotion of β cell proliferation, based on the observation that Igf1r haploinsufficiency increases the severity of diabetes in mice lacking IRS-2 (13). While our data do not support this view, there are potential explanations for the discrepancy between our observations and those in Irs2–/–Igf1r+/– mice. First, the tissue-specific phenotype that we describe here is not necessarily directly comparable with the state of ubiquitous haploinsufficiency in Igf1r+/– heterozygotes carrying a null allele. Second, it is possible that IRS-2, in addition to mediating IGF signaling, mediates signaling by other growth factors, potentially acting in a strain-dependent manner. Finally, it remains to be seen whether IR could play a compensatory role in β cell proliferation in the absence of IGF1R, as suggested by the observation that not only Igf1r haploinsufficiency, but also Ir haploinsufficiency results in accelerated β cell failure in Irs2–/– mice (58). Combined ablation of both the Ir and Igf1r functions in β cells is required to address this question.
In contrast to the absence of a proliferative defect, β cells lacking IGF1R exhibit a profound decrease of insulin secretion in response to both glucose and arginine. This impairment is reminiscent of the secretory defect observed in Irs1–/– β cells (26). Although the decrease in glucose-induced insulin secretion could be accounted for partly by the observed decrease in GLUT-2 expression, the inability of IGF1R-deficient β cells to respond to arginine suggests that the defect lies in the exocytotic pathway. Since arginine promotes insulin release through direct membrane depolarization and Ca2+ influx (59, 60), our data indicate that IGF1R signaling could be involved in regulating insulin secretion at a step distal to that of ATP production and closure of ATP-sensitive K channels (61). These findings are consistent with the observation that insulin stimulates exocytosis through changes in intracellular Ca2+ stores in a PI 3-kinase–dependent fashion (19). In this regard, it is important to note that while some steps of the exocytotic process are stimulated by insulin signaling, others may be inhibited. It was shown, for example, that intracellular translocation of storage granules into the early releasable pool is inhibited by PI 3-kinase activation (27). By analogy, it can be hypothesized that the early stimulatory effect of IGF-1 on insulin secretion in cultured β cells, which is followed by a late phase of inhibition (62), is due to activation of different steps in the exocytotic process.
Expression profiling using microarray analysis showed that there are no detectable differences in the transcription of known genes involved in insulin exocytosis. However, IGF1R signaling could promote this process by several additional mechanisms. For example, IGF1 may directly stimulate Ca2+ entry into β cells, occurring through translocation of a recently described growth factor–regulated calcium-permeable channel belonging to the TRP family (63). Consistent with this hypothesis, there are reports suggesting that IGF-2 promotes exocytosis in β cells (64), while tyrosine kinase inhibitors apparently prevent insulin secretion in response to several secretagogues (18). Alternatively, IGF signaling may activate ATP-sensitive K channels in a PI 3-kinase–dependent manner, as already shown for insulin (15), or could impinge upon G protein–mediated exocytotic steps through one of the insulin/IGF-sensitive G protein subunits (65). Finally, an intriguing possibility is that IGFs regulate insulin secretion through an autocrine mechanism. This hypothesis has emerged from studies of mice lacking HNF-1α (the MODY3 gene homologue) or overexpressing dominant negative mutant forms of HNF-1α in β cells (66–68), which develop a defect similar to that described here for β cell–specific Igf1r knockouts. Interestingly, IGF-1 appears to be an HNF-1α target gene in β cells that is downregulated in mice expressing a dominant negative HNF-1α (69). Furthermore, the hypothesis mentioned above could explain why overexpression of another IGF1R ligand, IGF-2, in β cells of transgenic mice results in increased insulin secretion (38).
While this manuscript was under review, Kulkarni et al., using different strains of Cre-producing and Cre-responding mice, reported the generation of mice lacking IGF1R in β cells (70). Their findings are in agreement with ours, except that they did not study the response of purified islets to arginine, but reported normal arginine-induced insulin secretion in vivo. Interestingly, both we and Kulkarni report an increase in basal insulin values, although in our study the values did not reach statistical significance.
The similarities between the phenotype described here and those of Ir and Irs1 knockouts indicate that IGF1R could have overlapping functions with those of Ir and could be signaling through IRS-1 in β cells. The identification of IGF-1–dependent mechanisms of exocytosis potentially could provide novel targets for therapeutic intervention in β cell dysfunction.
-
Acknowlegments
This work was supported by Juvenile Diabetes Research Foundation grant 2000-893 to D. Accili and A. Efstratiadis. We thank members of the Accili and Efstratiadis laboratories for helpful discussions.
-
Footnotes
Conflict of interest: No conflict of interest has been declared.
Nonstandard abbreviations used: phosphoinositide 3-kinase (PI 3-kinase); insulin receptor substrate (IRS); type 1 IGF receptor (IGF1R); insulin receptor (IR); 5-bromo-2-deoxyuridine (BrdU); embryonic day (E).
-
References
- Matschinsky, FM. Banting Lecture 1995. A lesson in metabolic regulation inspired by the glucokinase glucose sensor paradigm. Diabetes 1996. 45:223-241.
- Bonner-Weir, S. Regulation of pancreatic beta-cell mass in vivo. Recent Prog Horm Res 1994. 49:91-104. View this article via: PubMed Google Scholar
- Bonner-Weir, S. Life and death of the pancreatic beta cells. Trends Endocrinol Metab 2000. 11:375-378.
- Shaw, J, Latimer, E. Regeneration of pancreatic tissue from the transplanted pancreatic duct in the dog. Am J Physiol 1925. 76:49-53.
- Bendayan, M. Presence of endocrine cells in pancreatic ducts. Pancreas 1987. 2:393-397. View this article via: PubMed Google Scholar
- Bertelli, E, Regoli, M, Orazioli, D, Bendayan, M. Association between islets of Langerhans and pancreatic ductal system in adult rat. Where endocrine and exocrine meet together? Diabetologia 2001. 44:575-584.
- Bonner-Weir, S, et al. In vitro cultivation of human islets from expanded ductal tissue. Proc Natl Acad Sci USA 2000. 97:7999-8004.
- Bouwens, L, Pipeleers, DG. Extra-insular beta cells associated with ductules are frequent in adult human pancreas. Diabetologia 1998. 41:629-633.
- Hart, AW, Baeza, N, Apelqvist, A, Edlund, H. Attenuation of FGF signalling in mouse beta-cells leads to diabetes. Nature 2000. 408:864-868.
- Garcia-Ocana, , et al. Hepatocyte growth factor overexpression in the islet of transgenic mice increases beta cell proliferation, enhances islet mass, and induces mild hypoglycemia. J Biol Chem 2000. 275:1226-1232.
- Vasavada, RC, et al. Targeted expression of placental lactogen in the beta cells of transgenic mice results in beta cell proliferation, islet mass augmentation, and hypoglycemia. J Biol Chem 2000. 275:15399-15406.
- Rhodes, CJ. IGF-I and GH post-receptor signaling mechanisms for pancreatic beta-cell replication. J Mol Endocrinol 2000. 24:303-311.
- Withers, DJ, et al. Irs-2 coordinates Igf-1 receptor-mediated beta-cell development and peripheral insulin signalling. Nat Genet 1999. 23:32-40. View this article via: PubMed Google Scholar
- Hugl, SR, White, MF, Rhodes, CJ. Insulin-like growth factor I (IGF-I)-stimulated pancreatic beta-cell growth is glucose-dependent. Synergistic activation of insulin receptor substrate-mediated signal transduction pathways by glucose and IGF-I in INS-1 cells. J Biol Chem 1998. 273:17771-17779.
- Khan, FA, Goforth, PB, Zhang, M, Satin, LS. Insulin activates ATP-sensitive K(+) channels in pancreatic beta-cells through a phosphatidylinositol 3-kinase-dependent pathway. Diabetes 2001. 50:2192-2198.
- Leibiger, IB, Leibiger, B, Moede, T, Berggren, PO. Exocytosis of insulin promotes insulin gene transcription via the insulin receptor/PI 3-kinase/p70 s6 kinase and CaM kinase pathways. Mol Cell 1998. 1:933-938.
- Leibiger, B, et al. Selective insulin signaling through a and b insulin receptors regulates transcription of insulin and glucokinase genes in pancreatic beta cells. Mol Cell 2001. 7:559-570.
- Persaud, SJ, Harris, TE, Burns, CJ, Jones, PM. Tyrosine kinases play a permissive role in glucose-induced insulin secretion from adult rat islets. J Mol Endocrinol 1999. 22:19-28.
- Aspinwall, CA, et al. Roles of insulin receptor substrate-1, phosphatidylinositol 3-kinase, and release of intracellular Ca2+ stores in insulin-stimulated insulin secretion in beta-cells. J Biol Chem 2000. 275:22331-22338.
- Withers, DJ, et al. Disruption of IRS-2 causes type 2 diabetes in mice. Nature 1998. 391:900-904.
- Kubota, N, et al. Disruption of insulin receptor substrate 2 causes type 2 diabetes because of liver insulin resistance and lack of compensatory beta-cell hyperplasia. Diabetes 2000. 49:1880-1889.
- Pende, M, et al. Hypoinsulinaemia, glucose intolerance and diminished beta-cell size in S6K1-deficient mice. Nature 2000. 408:994-997.
- Tuttle, RL, et al. Regulation of pancreatic beta-cell growth and survival by the serine/threonine protein kinase Akt1/PKBalpha. Nat Med 2001. 7:1133-1137.
- Bernal-Mizrachi, E, Wen, W, Stahlhut, S, Welling, C, Permutt, M. Transgenic mice expressing a constitutively active Akt1/PKBα in pancreatic islet β-cells exhibit striking hypertrophy, hyperplasia and hyperinsulinemia. J Clin Invest 2001. 108:1631-1638.
- Kulkarni, RN, et al. Tissue-specific knockout of the insulin receptor in pancreatic beta cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell 1999. 96:329-339.
- Kulkarni, RN, et al. Altered function of insulin receptor substrate-1-deficient mouse islets and cultured beta-cell lines. J Clin Invest 1999. 104:R69-R75.
- Eto, K, et al. Phosphatidylinositol 3-kinase suppresses glucose-stimulated insulin secretion by affecting post-cytosolic [Ca(2+)] elevation signals. Diabetes 2002. 51:87-97.
- Zawalich, WS, Zawalich, KC. A link between insulin resistance and hyperinsulinemia: inhibitors of phosphatidylinositol 3-kinase augment glucose-induced insulin secretion from islets of lean, but not obese, rats. Endocrinology 2000. 141:3287-3295.
- Zhang, Q, Berggren, PO, Larsson, O, Hall, K, Tally, M. Insulin-like growth factor II inhibits glucose-induced insulin exocytosis. Biochem Biophys Res Commun 1998. 243:117-121.
- Zhao, AZ, Zhao, H, Teague, J, Fujimoto, W, Beavo, JA. Attenuation of insulin secretion by insulin-like growth factor 1 is mediated through activation of phosphodiesterase 3B. Proc Natl Acad Sci USA 1997. 94:3223-3228.
- Leahy, JL, Vandekerkhove, KM. Insulin-like growth factor-I at physiological concentrations is a potent inhibitor of insulin secretion. Endocrinology 1990. 126:1593-1598. View this article via: PubMed Google Scholar
- Petrik, J, et al. A low protein diet alters the balance of islet cell replication and apoptosis in the fetal and neonatal rat and is associated with a reduced pancreatic expression of insulin-like growth factor-II. Endocrinology 1999. 140:4861-4873.
- Hill, DJ, Hogg, J, Petrik, J, Arany, E, Han, VK. Cellular distribution and ontogeny of insulin-like growth factors (IGFs) and IGF binding protein messenger RNAs and peptides in developing rat pancreas. J Endocrinol 1999. 160:305-317.
- Hoog, A, et al. Increased amounts of a high molecular weight insulin-like growth factor II (IGF-II) peptide and IGF-II messenger ribonucleic acid in pancreatic islets of diabetic Goto-Kakizaki rats. Endocrinology 1996. 137:2415-2423.
- Portela-Gomes, GM, Hoog, A. Insulin-like growth factor II in human fetal pancreas and its co-localization with the major islet hormones: comparison with adult pancreas. J Endocrinol 2000. 165:245-251.
- Scaglia, L, Cahill, CJ, Finegood, DT, Bonner-Weir, S. Apoptosis participates in the remodeling of the endocrine pancreas in the neonatal rat. Endocrinology 1997. 138:1736-1741.
- Petrik, J, Arany, E, McDonald, TJ, Hill, DJ. Apoptosis in the pancreatic islet cells of the neonatal rat is associated with a reduced expression of insulin-like growth factor II that may act as a survival factor. Endocrinology 1998. 139:2994-3004.
- Devedjian, JC, et al. Transgenic mice overexpressing insulin-like growth factor-II in beta cells develop type 2 diabetes. J Clin Invest 2000. 105:731-740.
- Rossetti, L, et al. Hepatic overexpression of insulin-like growth factor-II in adulthood increases basal and insulin-stimulated glucose disposal in conscious mice. J Biol Chem 1996. 271:203-208.
- Petrik, J, et al. Overexpression of insulin-like growth factor-II in transgenic mice is associated with pancreatic islet cell hyperplasia. Endocrinology 1999. 140:2353-2363.
- Liu, JP, Baker, J, Perkins, AS, Robertson, EJ, Efstratiadis, A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell 1993. 75:59-72. View this article via: PubMed Google Scholar
- Dietrich, P, Dragatsis, I, Xuan, S, Zeitlin, S, Efstratiadis, A. Conditional mutagenesis in mice with heat shock promoter-driven cre transgenes. Mamm Genome 2000. 11:196-205.
- Herrera, PL. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development 2000. 127:2317-2322. View this article via: PubMed Google Scholar
- Di Cola, G, Cool, MH, Accili, D. Hypoglycemic effect of insulin-like growth factor-1 in mice lacking insulin receptors. J Clin Invest 1997. 99:2538-2544.
- Kido, Y, et al. Tissue-specific insulin resistance in mice with combined mutations of insulin receptor, IRS-1 and IRS-2. J Clin Invest 2000. 105:199-205.
- Kitamura, T, et al. Preserved pancreatic beta-cell development and function in mice lacking the insulin receptor-related receptor. Mol Cell Biol 2001. 21:5624-5630.
- Klein, U, et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J. Exp Med 2001. 194:1625-1638. View this article via: CrossRef Google Scholar
- Aronoff, SL, Bennett, PH, Gorden, P, Rushforth, N, Miller, M. Unexplained hyperinsulinemia in normal and “prediabetic” Pima Indians compared with normal Caucasians. An example of racial differences in insulin secretion. Diabetes 1977. 26:827-840.
- Lillioja, S, et al. Exaggerated early insulin release and insulin resistance in a diabetes-prone population: a metabolic comparison of Pima Indians and Caucasians. J Clin Endocrinol Metab 1991. 73:866-876. View this article via: PubMed Google Scholar
- O’Rahilly, S, Turner, RC, Matthews, DR. Impaired pulsatile secretion of insulin in relatives of patients with non-insulin-dependent diabetes. N Engl J Med 1988. 318:1225-1230. View this article via: PubMed Google Scholar
- Herman, WH, et al. Abnormal insulin secretion, not insulin resistance, is the genetic or primary defect of MODY in the RW pedigree. Diabetes 1994. 43:40-46.
- Byrne, MM, et al. Altered insulin secretory responses to glucose in diabetic and nondiabetic subjects with mutations in the diabetes susceptibility gene MODY3 on chromosome 12. Diabetes 1996. 45:1503-1510.
- Bell, GI, Polonsky, KS. Diabetes mellitus and genetically programmed defects in beta-cell function. Nature 2001. 414:788-791.
- Accili, D, Kido, Y, Nakae, J, Lauro, D, Park, B-C. Genetics of type 2 diabetes: insights from targeted mouse mutants. Curr Mol Med 2001. 1:9-23.
- Flier, SN, Kulkarni, RN, Kahn, CR. Evidence for a circulating islet cell growth factor in insulin-resistant states. Proc Natl Acad Sci USA 2001. 98:7475-7480.
- Bonner-Weir, S. Perspective: postnatal pancreatic beta cell growth. Endocrinology 2000. 141:1926-1929.
- Accili, D. A kinase in the life of the beta cell. J Clin Invest 2001. 108:1575-1576.
- Kitamura, T, et al. The transcription factor FKHR regulates pancreatic beta cell survival. Diabetes 2001. 50(Suppl. 2):A345.
- Henquin, J. 1992. The biophysical events involved in the stimulation of insulin release by arginine. In Guanidino compounds in biology and medicine. P. DeDeyen, B. Marescau, V. Stalon, and I. Qureshi, editors. John Wiley & Co. New York, New York, USA. 109–116.
- Charles, S, Tamagawa, T, Henquin, JC. A single mechanism for the stimulation of insulin release and 86Rb+ efflux from rat islets by cationic amino acids. Biochem J 1982. 208:301-308. View this article via: PubMed Google Scholar
- Henquin, JC. Triggering and amplifying pathways of regulation of insulin secretion by glucose. Diabetes 2000. 49:1751-1760.
- Hill, DJ, Sedran, RJ, Brenner, SL, McDonald, TJ. IGF-I has a dual effect on insulin release from isolated, perifused adult rat islets of Langerhans. J Endocrinol 1997. 153:15-25.
- Kanzaki, M, et al. Translocation of a calcium-permeable cation channel induced by insulin-like growth factor-I. Nat Cell Biol 1999. 1:165-170.
- Zhang, Q, et al. Insulin-like growth factor II signaling through the insulin-like growth factor II/mannose-6-phosphate receptor promotes exocytosis in insulin-secreting cells. Proc Natl Acad Sci USA 1997. 94:6232-6237.
- Lang, J. Molecular mechanisms and regulation of insulin exocytosis as a paradigm of endocrine secretion. Eur J Biochem 1999. 259:3-17.
- Hagenfeldt-Johansson, KA, et al. Beta-cell-targeted expression of a dominant-negative hepatocyte nuclear factor-1 alpha induces a maturity-onset diabetes of the young (MODY)3-like phenotype in transgenic mice. Endocrinology 2001. 142:5311-5320.
- Yamagata, K, et al. Overexpression of dominant-negative mutant hepatocyte nuclear factor-1alpha in pancreatic beta-cells causes abnormal islet architecture with decreased expression of E-Cadherin, reduced beta-cell proliferation, and diabetes. Diabetes 2002. 51:114-123.
- Pontoglio, M, et al. Defective insulin secretion in hepatocyte nuclear factor 1alpha-deficient mice. J Clin Invest 1998. 101:2215-2222.
- Yang, Q, et al. Hepatocyte nuclear factor-1alpha modulates pancreatic beta-cell growth by regulating the expression of insulin-like growth factor-1 in INS-1 cells. Diabetes 2002. 51:1785-1792.
- Kulkarni, RN, et al. Beta-cell-specific deletion of the Igf1 receptor leads to hyperinsulinemia and glucose intolerance but does not alter beta-cell mass. Nat Genet 2002. 31:111-115. View this article via: PubMed Google Scholar
-
Version history
- Version 1 (October 1, 2002): No description



Copyright © 2026 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)










