Advertisement
Research ArticleCardiologyCell biology Free access | 10.1172/JCI134682
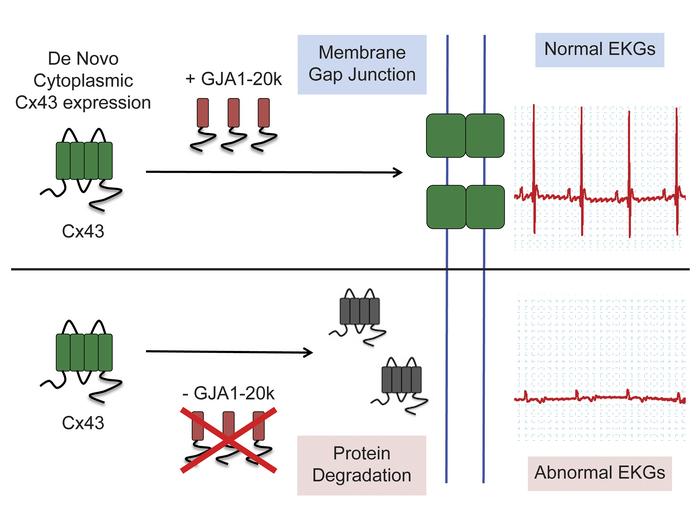
Auxiliary trafficking subunit GJA1-20k protects connexin-43 from degradation and limits ventricular arrhythmias
Shaohua Xiao,1,2 Daisuke Shimura,1 Rachel Baum,1,3 Diana M. Hernandez,3 Sosse Agvanian,3 Yoshiko Nagaoka,3,4 Makoto Katsumata,3 Paul D. Lampe,5 Andre G. Kleber,6 TingTing Hong,1,3 and Robin M. Shaw1
1Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, Salt Lake City, Utah, USA.
2Department of Neurology, UCLA, Los Angeles, California, USA.
3Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, California, USA.
4Department of Pathology, University of Alabama at Birmingham, Birmingham, Alabama, USA.
5Translational Research Program, Public Health Sciences and Human Biology Divisions, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
6Department of Pathology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Robin M. Shaw, Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, 95 South 2000 East, Suite 143, Salt Lake City, Utah 84112, USA. Phone: 801.581.8183; Email: Robin.Shaw@hsc.utah.edu.
Authorship note: SX and DS contributed equally to this work.
Find articles by Xiao, S. in: PubMed | Google Scholar
1Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, Salt Lake City, Utah, USA.
2Department of Neurology, UCLA, Los Angeles, California, USA.
3Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, California, USA.
4Department of Pathology, University of Alabama at Birmingham, Birmingham, Alabama, USA.
5Translational Research Program, Public Health Sciences and Human Biology Divisions, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
6Department of Pathology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Robin M. Shaw, Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, 95 South 2000 East, Suite 143, Salt Lake City, Utah 84112, USA. Phone: 801.581.8183; Email: Robin.Shaw@hsc.utah.edu.
Authorship note: SX and DS contributed equally to this work.
Find articles by
Shimura, D.
in:
PubMed
|
Google Scholar
|

1Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, Salt Lake City, Utah, USA.
2Department of Neurology, UCLA, Los Angeles, California, USA.
3Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, California, USA.
4Department of Pathology, University of Alabama at Birmingham, Birmingham, Alabama, USA.
5Translational Research Program, Public Health Sciences and Human Biology Divisions, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
6Department of Pathology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Robin M. Shaw, Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, 95 South 2000 East, Suite 143, Salt Lake City, Utah 84112, USA. Phone: 801.581.8183; Email: Robin.Shaw@hsc.utah.edu.
Authorship note: SX and DS contributed equally to this work.
Find articles by Baum, R. in: PubMed | Google Scholar
1Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, Salt Lake City, Utah, USA.
2Department of Neurology, UCLA, Los Angeles, California, USA.
3Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, California, USA.
4Department of Pathology, University of Alabama at Birmingham, Birmingham, Alabama, USA.
5Translational Research Program, Public Health Sciences and Human Biology Divisions, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
6Department of Pathology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Robin M. Shaw, Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, 95 South 2000 East, Suite 143, Salt Lake City, Utah 84112, USA. Phone: 801.581.8183; Email: Robin.Shaw@hsc.utah.edu.
Authorship note: SX and DS contributed equally to this work.
Find articles by Hernandez, D. in: PubMed | Google Scholar
1Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, Salt Lake City, Utah, USA.
2Department of Neurology, UCLA, Los Angeles, California, USA.
3Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, California, USA.
4Department of Pathology, University of Alabama at Birmingham, Birmingham, Alabama, USA.
5Translational Research Program, Public Health Sciences and Human Biology Divisions, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
6Department of Pathology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Robin M. Shaw, Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, 95 South 2000 East, Suite 143, Salt Lake City, Utah 84112, USA. Phone: 801.581.8183; Email: Robin.Shaw@hsc.utah.edu.
Authorship note: SX and DS contributed equally to this work.
Find articles by Agvanian, S. in: PubMed | Google Scholar
1Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, Salt Lake City, Utah, USA.
2Department of Neurology, UCLA, Los Angeles, California, USA.
3Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, California, USA.
4Department of Pathology, University of Alabama at Birmingham, Birmingham, Alabama, USA.
5Translational Research Program, Public Health Sciences and Human Biology Divisions, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
6Department of Pathology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Robin M. Shaw, Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, 95 South 2000 East, Suite 143, Salt Lake City, Utah 84112, USA. Phone: 801.581.8183; Email: Robin.Shaw@hsc.utah.edu.
Authorship note: SX and DS contributed equally to this work.
Find articles by Nagaoka, Y. in: PubMed | Google Scholar
1Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, Salt Lake City, Utah, USA.
2Department of Neurology, UCLA, Los Angeles, California, USA.
3Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, California, USA.
4Department of Pathology, University of Alabama at Birmingham, Birmingham, Alabama, USA.
5Translational Research Program, Public Health Sciences and Human Biology Divisions, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
6Department of Pathology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Robin M. Shaw, Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, 95 South 2000 East, Suite 143, Salt Lake City, Utah 84112, USA. Phone: 801.581.8183; Email: Robin.Shaw@hsc.utah.edu.
Authorship note: SX and DS contributed equally to this work.
Find articles by
Katsumata, M.
in:
PubMed
|
Google Scholar
|

1Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, Salt Lake City, Utah, USA.
2Department of Neurology, UCLA, Los Angeles, California, USA.
3Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, California, USA.
4Department of Pathology, University of Alabama at Birmingham, Birmingham, Alabama, USA.
5Translational Research Program, Public Health Sciences and Human Biology Divisions, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
6Department of Pathology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Robin M. Shaw, Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, 95 South 2000 East, Suite 143, Salt Lake City, Utah 84112, USA. Phone: 801.581.8183; Email: Robin.Shaw@hsc.utah.edu.
Authorship note: SX and DS contributed equally to this work.
Find articles by
Lampe, P.
in:
PubMed
|
Google Scholar
|

1Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, Salt Lake City, Utah, USA.
2Department of Neurology, UCLA, Los Angeles, California, USA.
3Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, California, USA.
4Department of Pathology, University of Alabama at Birmingham, Birmingham, Alabama, USA.
5Translational Research Program, Public Health Sciences and Human Biology Divisions, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
6Department of Pathology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Robin M. Shaw, Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, 95 South 2000 East, Suite 143, Salt Lake City, Utah 84112, USA. Phone: 801.581.8183; Email: Robin.Shaw@hsc.utah.edu.
Authorship note: SX and DS contributed equally to this work.
Find articles by Kleber, A. in: PubMed | Google Scholar
1Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, Salt Lake City, Utah, USA.
2Department of Neurology, UCLA, Los Angeles, California, USA.
3Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, California, USA.
4Department of Pathology, University of Alabama at Birmingham, Birmingham, Alabama, USA.
5Translational Research Program, Public Health Sciences and Human Biology Divisions, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
6Department of Pathology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Robin M. Shaw, Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, 95 South 2000 East, Suite 143, Salt Lake City, Utah 84112, USA. Phone: 801.581.8183; Email: Robin.Shaw@hsc.utah.edu.
Authorship note: SX and DS contributed equally to this work.
Find articles by
Hong, T.
in:
PubMed
|
Google Scholar
|

1Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, Salt Lake City, Utah, USA.
2Department of Neurology, UCLA, Los Angeles, California, USA.
3Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, California, USA.
4Department of Pathology, University of Alabama at Birmingham, Birmingham, Alabama, USA.
5Translational Research Program, Public Health Sciences and Human Biology Divisions, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
6Department of Pathology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Robin M. Shaw, Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, 95 South 2000 East, Suite 143, Salt Lake City, Utah 84112, USA. Phone: 801.581.8183; Email: Robin.Shaw@hsc.utah.edu.
Authorship note: SX and DS contributed equally to this work.
Find articles by
Shaw, R.
in:
PubMed
|
Google Scholar
|

Authorship note: SX and DS contributed equally to this work.
Published June 11, 2020 - More info
J Clin Invest. 2020;130(9):4858–4870. https://doi.org/10.1172/JCI134682.
© 2020 American Society for Clinical Investigation
Received: November 4, 2019; Accepted: June 3, 2020
-
Abstract
Connexin-43 (Cx43) gap junctions provide intercellular coupling, which ensures rapid action potential propagation and synchronized heart contraction. Alterations in Cx43 localization and reductions in gap junction coupling occur in failing hearts, contributing to ventricular arrhythmias and sudden cardiac death. Recent reports have found that an internally translated Cx43 isoform, GJA1-20k, is an auxiliary subunit for the trafficking of Cx43 in heterologous expression systems. Here, we have created a mouse model by using CRISPR technology to mutate a single internal translation initiation site in Cx43 (M213L mutation), which generates full-length Cx43, but not GJA1-20k. We found that GJA1M213L/M213L mice had severely abnormal electrocardiograms despite preserved contractile function, reduced total Cx43, and reduced gap junctions, and they died suddenly at 2 to 4 weeks of age. Heterozygous GJA1M213L/WT mice survived to adulthood with increased ventricular ectopy. Biochemical experiments indicated that cytoplasmic Cx43 had a half-life that was 50% shorter than membrane-associated Cx43. Without GJA1-20k, poorly trafficked Cx43 was degraded. The data support that GJA1-20k, an endogenous entity translated independently of Cx43, is critical for Cx43 gap junction trafficking, maintenance of Cx43 protein, and normal electrical function of the mammalian heart.
-
Introduction
Intercellular communication between cardiomyocytes via connexin-43 (Cx43) gap junctions enables the rapid spread of electrical signals and synchronous ventricular contraction. Oligomerization of 6 Cx43 molecules gives rise to a connexon or hemichannel, which binds to a connexon from the adjacent cell to form a full gap junction channel. Cx43 gap junction channels are primarily localized to the intercalated discs (ICDs) at the longitudinal ends of cardiomyocytes (1). Loss of Cx43 gap junction coupling contributes to abnormal propagation of electrical impulse, arrhythmias, and sudden cardiac death (2, 3). Similar to other cardiac ion channels, Cx43 has a short half-life, ranging from 1 to 3 hours (4, 5). Given the importance of properly localized Cx43 at gap junctions and its rapid turnover, forward trafficking and localization of Cx43 are under intricate regulation that is only beginning to be understood.
In heterologous expression systems, it has been identified that a short isoform of Cx43, GJA1-20k, serves as a chaperone for the assembly and trafficking of full-length Cx43 gap junction channels (5–8). The GJA1-20k isoform is produced as a result of alternative translation of mRNA, by which AUG codons (typically coding for methionine) within the coding region of the GJA1 mRNA function as ribosomal start sites to initiate translation. Thus, protein formation can be initiated downstream of the canonical translation start site, generating N-terminal truncated protein isoforms (6). GJA1-20k is the most abundant internally translated Cx43 isoform endogenously expressed in the heart (6). In isolated cells, full-length Cx43 introduced heterologously by plasmids, whose corresponding mRNA is unable to generate GJA1-20k, fail to be delivered to cell-cell junctions; the addition of exogenously expressed GJA1-20k rescues the trafficking defects of full-length protein (6). The facilitation of GJA1-20k in forward trafficking is related to its interaction with the cytoskeleton, which serves as the membrane protein delivery apparatus. GJA1-20k organizes the actin and microtubule cytoskeleton for targeted delivery of Cx43 hemichannels to cell-cell borders (7). Despite knowledge of GJA1-20k participation in Cx43 trafficking, the life cycle of Cx43 channels in adult heart cells remains poorly understood. It is not known whether channels in the plasma membrane behave differently than those not yet delivered. If channels cannot be trafficked, can they remain available in the cardiomyocyte or are they degraded?
The questions regarding Cx43 movement and life cycle have immediate relevance to cardiac health. If Cx43 channels are limited in their ability to traffic to the cardiac ICD, do they remain available for ultra-rapid delivery after a stimulus, or would new channels need to be generated before enhancement of gap junction coupling? How prepared are mammalian hearts for a sudden need for increased intercellular coupling? We have found that actin stabilization and pretreatment with GJA1-20k can preserve coupling in adult hearts subsequently subjected to ischemic stress (7, 9). Pretreatment with GJA1-20k is also cardioprotective (10). Understanding the role of GJA1-20k in regulating Cx43 will guide therapies for gap junction rescue, such as helping to determine whether therapeutic interventions could be immediately effective after the start of ischemia or require initiation of treatment before the onset of an ischemic period.
To explore the movement of Cx43 in vivo and in adult cardiomyocytes, we generated a mouse model deprived of GJA1-20k using the CRISPR homology directed repair (HDR) technology to mutate the single internal methionine initiation codon for GJA1-20k (GJA1-M213L mutation, corresponding to substitution of AUG by UUA in mRNA). This mutation was in a highly conserved region of Cx43 DNA that did not affect production of full-length Cx43, but removed endogenous generation of GJA1-20k. The results were that without its smaller trafficking partner, full-length Cx43 did not reach the ICD, and mice without GJA1-20k formation died from apparent sudden cardiac death. Surprisingly, we found that poorly trafficked Cx43 was degraded rather than stored in cytoplasmic reserves. The stability of ICD-associated Cx43 was much higher than that of cytoplasmic Cx43.
-
Results
GJA1M213L/M213L mice die suddenly. To explore perturbation of Cx43 trafficking in vivo, we created a mouse model that reduced the ability of GJA1-20k to be formed by replacing the internal ribosomal translation start site, the AUG-encoding amino acid 213 (methionine 213) of the mouse Cx43, to UUA (encoding leucine, L). We used CRISPR-HDR technology to edit the genomic sequence at the target location (Figure 1A and Supplemental Figure 1; supplemental material available online with this article;http://doi.org/10.1172/JCI134682DS1 The sequence around the target site in the WT allele was sensitive to NlaIII restriction digestion, whereas that in the M213L mutated allele was resistant. Therefore, PCR amplification and NlaIII digestion produced 2, 3, and 1 DNA fragments in the genotyping assay of WT, heterozygotes (HT), and homozygotes (HM), respectively (Figure 1B). The M213L mutation did not significantly change the expression level of the GJA1 mRNA (Figure 1C). However, the expression of the GJA1-20k protein was almost completely removed in mice homozygous for the M213L mutation compared with the WT and HT littermates (Figure 1, D and E). These observations are consistent with previous studies of heterologous expression systems, demonstrating that the GJA1-20k protein is produced as a result of alternative translation initiation (6, 11, 12). Taken together, the level of endogenous GJA1-20k protein was effectively diminished in mice carrying the M213L mutation. We thus refer to this mouse model as a GJA1M213L/M213L for the homozygous mutant and GJA1M213L/WT for the heterozygous mutant.
 Figure 1
Figure 1Generation of the M213L mouse model and survival of the mutant mice. (A) Schematic diagram showing the site-directed mutation of ATG, encoding M213, to TTA, encoding an L, at the GJA1 genomic locus using CRISPR technology. HDR: homology directed repair. (B) Representative genotyping results confirmed presence of the M213L mutation in 0, 1, and both copies of the GJA1 gene in WT mice, heterozygous (HT), and homozygous (HM) for M213L, respectively. (C) Quantification of the GJA1 mRNA expression in the hearts of WT, HT, and HM mice. One-way ANOVA, with n = 4 in each group. Data represent mean ± SEM. (D) Representative Western blot showing the expression of GJA1-20k protein in cardiac ventricles of WT, HT, and HM mice (2 to 3 weeks old) after immunoprecipitation. n = 3 experiments. (E) Quantification of GJA1-20k protein expression in cardiac ventricles of 2- to 3-week-old mice among genotypes. Kruskal-Wallis test, followed by Dunn’s multiple-comparisons test, with n = 3 in each group. Data represent mean ± SEM. (F) Kaplan-Meier survival curve of the WT (n = 18, black), HT (n = 36, green), and HM (n = 16, red) mice. Survival curves were compared using log-rank (Mantel-Cox) test.
It should be emphasized that although our use of the terms “homozygous,” “heterozygous,” and “mutation” is technically correct, these terms typically refer to mutations in genes that are associated with an absence or change in the corresponding full-length protein. In the case of our M213L mutation, we changed an internal ribosomal start site. Even for the homozygous GJA1M213L/M213L mouse, full-length Cx43 mRNA and protein were still generated, yet there was decreased internal ribosomal translation and decreased production of the smaller GJA1-20k protein. As a result, any changes to Cx43 expression with an M213L mutation were therefore secondary to posttranslational Cx43 movements and processing in the presence of unchanged primary synthesis yet reduced GJA1-20k.
The GJA1M213L/M213L mice grew similarly to the WT and heterozygous littermates for 2 weeks, without a difference in gross physical appearance (Figure 2, A–C). However, they died suddenly around 2–4 weeks old, with the median lifespan of 18 days (Figure 1F). In addition, young GJA1M213L/M213L mice did not show abnormalities in the right ventricular outflow tract (RVOT) or in other cardiac regions, although it was previously reported that a mouse lacking full-length Cx43 developed RVOT outflow tract obstruction and death at birth (13). The survival to 2–4 weeks (Figure 1F) and normal physical development (Figure 2, A–C) argue against RVOT outflow tract obstruction. We also performed echocardiography on these mice at 2 to 3 weeks of age (Figure 2D) and found normal left ventricular (LV) and right ventricular (RV) size and function, indicating preserved hemodynamics despite a loss of GJA1-20k. Full echocardiographic parameters of the young mice are presented in Table 1, which indicates that heart rate, as well as all 13 other structural and functional parameters of the heart, were not changed among WT, GJA1M213L/WT, and GJA1M213L/M213L mice. The only significant difference was the long axis fractional shortening, which was mildly reduced in GJA1M213L/M213L mice (12.96% ± 1.17%) compared with WT (19.11% ± 1.07%) and GJA1M213L/WT (18.78% ± 1.07%) littermates (Table 1). Overall ejection fraction and LV systolic and diastolic volumes were unchanged (Table 1).
 Figure 2
Figure 2Morphology of the hearts and gross physical parameters of young (2–3 weeks) M213L mice. (A and B) Quantification of body weight (A) and body length (B) of WT, HT, and HM mice. (C) A representative image of WT and HM mice. (D) Representative long axis echocardiograph images of young WT, HT, and HM mice. The lower panels indicate the trace images of heart wall. LV: left ventricle; RV: right ventricle. One-way ANOVA test. The number of mice (n) in each group is indicated within bars or Table 1. Data represent mean ± SEM.
Although GJA1M213L/M213L mice died young, we were able to measure echocardiographic parameters to assess the structure and hemodynamics of the GJA1M213L/WT and WT adult mouse hearts. In mice 9–10 weeks old, only LV end diastolic volume was significantly yet mildly reduced in GJA1M213L/WT (43.61 ± 2.26 μL) compared with WT littermates (55.52 ± 3.65 μL). All 13 other structural and functional parameters were unchanged (Supplemental Table 1). The echocardiographic data were supportive, in that structural and mechanical parameters were not significantly affected by the M213L mutation, further indicating that impaired mechanical cardiac function cannot explain the early sudden death phenotype of GJA1M213L/M213L mice.
GJA1M213L/M213L and GJA1M213L/WT mice have abnormal cardiac electrical excitation. We explored differences in electrical function as a potential mechanism of the sudden death of GJA1M213L/M213L mice, as illustrated in Figure 3. ECGs were recorded to examine cardiac whole-heart excitation in anesthetized GJA1M213L/M213L and GJA1M213L/WT mice. In young mice (2–3 weeks of age), a paucity of GJA1-20k resulted in a greatly reduced R wave amplitude. WT (0.8740 ± 0.05465 mV, mean ± SEM) R wave amplitude was reduced by 16% in GJA1M213L/WT mice (0.7360 ± 0.03750 mV), and by a remarkable 92% in GJA1M213L/M213L mice (0.06773 ± 0.01391 mV) (Figure 3, A–C). Duration of the Q, R, and S wave (QRS) complexes was tripled in the GJA1M213L/M213L mice (38.57 ± 5.98 ms) compared with WT mice (12.97 ± 0.57 ms) (Figure 3, A–C), indicating impaired ventricular excitation, probably as a result of global propagation slowing (14). Next, we used implanted telemetry to monitor ECGs of conscious and active adult (6–7 months old) WT and GJA1M213L/WT mice. The HT mice developed significantly more premature ventricular contractions (PVCs) than WT mice (Figure 3D). The average number of ectopic incidents per hour (including single PVCs and 2–4 consecutive PVCs) was 141.7 ± 23.16 (mean ± SEM) in GJA1M213L/WT mice versus 23.40 ± 8.45 in WT mice (Figure 3E). The decrease in R wave amplitude, lengthening of QRS complex duration, and increase in frequency of PVCs indicate that, even in GJA1M213L/WT animals, cardiac electric excitation was impaired when GJA1-20k expression was reduced.
 Figure 3
Figure 3ECGs of young (14–22 days) and adult (6–7 months) M213L mice. (A) Representative ECG traces of anesthetized young WT, HT, and HM mice. Quantification of the amplitude of R waves (B) and QRS duration (C) in young WT, HT, and HM mice. Kruskal-Wallis test, followed by Dunn’s multiple-comparisons test. (D) Examples of 1, 2, and 3 consecutive PVCs occurred in adult HT mice during the dark cycle recorded by telemetry. (E) Quantification of the number of PVC incidents in adult WT and HT mice. Mann-Whitney test with 2-tailed P value. (C–E) Data represent mean ± SEM. *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001. The number of mice (n) in each group is indicated in parentheses or within bars.
Expression of full-length Cx43 at the ICDs is reduced in GJA1M213L/M213L mutant mouse hearts with GJA1-20k depletion. Since GJA1-20k has been shown to facilitate delivery of full-length Cx43 to cell-cell junctions in cultured cell lines and primary cardiomyocytes (6, 7) and reduced expression of Cx43 is associated with decreased voltage in QRS complexes (15–17), we examined the expression of Cx43 at the ICDs in GJA1M213L mutant mouse hearts, as depicted in Figure 4. Indeed, full-length Cx43 had minimum expression at the ICDs, in contrast to the expected localization at ICDs in WT hearts (Figure 4A). Quantification of the Cx43 signal at the ICDs, which were delineated by N-cadherin staining, revealed a 53.9% reduction in GJA1M213L/M213L (462.7 ± 20.71 units) versus WT (1004 ± 65.02 units) hearts (Figure 4B). In heterozygous GJA1M213L/WT cardiomyocytes, Cx43 was similar to WT hearts (Figure 4, A and B). As expected (18, 19), there was no difference in N-cadherin expression among WT, HT, and HM hearts (Supplemental Figure 2), which suggests that the impairment of Cx43 localization to ICDs may result from impaired forward trafficking of hemichannels to the otherwise intact disc (20). Total Cx43 protein in the ventricles was also explored by Western blot (Figure 4C) and quantified (Figure 4D). The reduction of Cx43 in adult myocardium of GJA1M213L/M213L mice was surprisingly severe (Figure 4D), despite unaltered levels of Cx43 mRNA (Figure 1C), attributing this Cx43 reduction to a posttranscriptional event. This is in line with an earlier finding of rescue of full-length Cx43 localization at the ICD by GJA1-20k alone even when Cx43 had a M213L mutation (6). Therefore, the loss of Cx43 in the GJA1M213L/M213L mice was a posttranslational effect on Cx43, reflecting increased protein degradation.
 Figure 4
Figure 4Localization and expression of full-length Cx43 and Nav1.5 in cardiac ventricles of young (2–3 weeks) M213L mice. (A and E) Representative average projected immunofluorescence images of N-cadherin (NCad), full-length (FL) Cx43 (A), and Nav1.5 (E), in cryosections of cardiac ventricles. Cx43 and Nav1.5 are shown in green, and NCad is shown in magenta. Colocalization is shown as white in the merged images. Scale bar: 15 μm. (B and F) Quantification of the immunofluorescence signal of Cx43 (FL) (B) and Nav1.5 (F) at the intercalated discs that are identified with NCad. ****P ≤ 0.0001, Kruskal-Wallis test, followed by Dunn’s multiple-comparisons test, n = 25, 21, and 22 images (B) and n = 34, 29, and 28 images (F) of WT, HT, and HM mice, respectively, with 6 mice in each group. Data represent mean ± SEM. (C and G) Representative Western blots showing the expression of Cx43 (FL) (C) and Nav1.5 (G) protein in the cardiac ventricles of WT, HT, and HM mice. n = 3 experiments. (D and H) Quantification of Cx43(FL) (D) and Nav1.5 (H) protein expression. The values were normalized to GAPDH expression. All graph data represent mean ± SEM. *P ≤ 0.05, Kruskal-Wallis test, followed by Dunn’s multiple-comparisons test, n = 3 mice in each group.
Cx43 gap junctions are responsible for the cell-to-cell spatial spread of electrotonic current in ventricular muscle. Severely reduced Cx43 gap junction localization to ICDs would cause delayed excitation, explaining the reduced QRS amplitudes with QRS notching and widening seen in Figure 3. After the membrane electrotonic downstream current has reached the threshold for sodium channel (Nav1.5) activation, the sodium current, as the main charge carrier, will produce the action potential upstroke and further downstream flow of electrotonic currents (21). This crucial complementary role of Cx43 and Nav1.5 prompted us to explore whether loss of GJA1-20k affects sodium channel localization at the ICD as well. In the GJA1M213L/M213L mice and GJA1M213L/WT mice, sodium channel localization did not appear to be affected (Figure 4, E and F). There was also no change in overall sodium channel protein (Figure 4, G and H). These results indicate that the GJA1-20k effect was not necessary for sodium channel trafficking.
M213L mutation affects full-length Cx43 forward delivery to cell-cell junctions. As described in Figure 4, the levels of full-length Cx43 protein were markedly reduced in the hearts of GJA1M213L/M213L mice compared with WT hearts, whereas the Cx43 expression level in GJA1M213L/WT mice was intermediate (Figure 4D). We explored whether the decrease of cellular Cx43 protein occurred in the cytoplasmic compartment of 2- to 3-week-old mouse ventricular cardiomyocytes or by loss of Cx43 at the ICD. Triton fractionation was performed to separate and quantify the soluble (cytoplasmic) fraction of Cx43 from the insoluble (ICD) fraction (ref. 18 and Figure 5). The results indicate that although the reduction of Cx43 at the ICD was striking in GJA1M213L/M213L mice (Figure 5C), the amount of poorly trafficked Cx43 in the cytoplasm was slightly increased, but still surprisingly close to that of WT mice. In Figure 5D we compare for the WT (left column) and GJA1M213L/M213L (right column) mice the relative contribution of ICD Cx43 (dark regions) and cytoplasmic Cx43 (light regions) to total Cx43 protein. In WT mice, there were roughly similar amounts of Cx43 at the ICD and in the cytoplasm. In GJA1M213L/M213L mice, the reduction in Cx43 protein was due to reduction in ICD Cx43, without a compensatory increase in cytoplasmic Cx43, implying that the Cx43 that was not trafficked to the ICD was degraded.
 Figure 5
Figure 5Cx43 expression in Triton-soluble (cytoplasmic) and -insoluble (junctional membrane) fractions in young (2–3 weeks) mouse hearts. (A) Representative Western blots showing the expression of Cx43 (FL), N-cadherin, and tubulin. (B and C) The quantification of graphs of Cx43 (FL) expression in soluble (B) and insoluble (C) fractions from WT and HM mouse hearts. The insoluble fraction of Cx43 (FL) was drastically decreased in HM. (D) The fold change of Cx43 (FL) in total (both soluble and insoluble). All graph data represent mean ± SEM. *P ≤ 0.05, **P ≤ 0.01 Mann-Whitney test with 2-tailed P value. n = 5 mice in each group.
The results of Figure 5 are supportive of a model that loss of GJA1-20k impaired forward trafficking of full-length Cx43 to the ICDs, which has already been established in cell lines and neonatal cardiomyocytes (6). However, we found that failure of Cx43 forward trafficking led to faster Cx43 protein degradation, suggesting that the pool of cytoplasmic Cx43 was degraded faster than the pool of Cx43 successfully trafficked to the ICD. It is theoretically possible that the M213L mutation itself compromises cytoplasmic protein stability. To explore whether the different degradation rates for cytoplasmic Cx43 versus ICD Cx43 could be due to instability induced by the M213L mutation, we used pulse-chase experiments to assess the protein stability of full-length mouse Cx43 with and without the M213L mutation (Figure 6). In HEK293 cells, we exogenously expressed either WT GJA1 or GJA1-M213L plasmids, both tagged with human influenza HA. As expected, the presence of the M213L mutation resulted in reduced translation of GJA1-20k, as indicated by the reduced levels of GJA1-20k protein (Supplemental Figure 3). Furthermore, the stability of full-length WT Cx43 and mutated Cx43-M213L was similar (Figure 6, A–C). In the Triton-soluble (cytoplasmic) fraction, the time constant tau (τ) of WT Cx43 and Cx43-M213L was similar, at 3.39 and 3.53 hours, respectively (Figure 6C). In fact, the M213L mutation resulted in a slightly longer time constant. In the Triton-insoluble fraction, the τ of WT Cx43 and Cx43-M213L was also similar, at 4.95 and 5.34 hours, respectively (Figure 6C). Even in the membrane fraction, the τ of Cx43-M213L was slightly longer than that of WT Cx43.
 Figure 6
Figure 6Stabilities of exogenous WT and M213L Cx43 protein in HEK cells. (A) Representative Western blots of the immunoprecipitated (IP), pulsed (blotted with streptavidin [STR]) and total (blotted with Cx43 antibody) exogenously expressed full-length Cx43 (FL) in Triton-soluble and -insoluble fractions at various time points during the chase period. n = 4 and 3 independent experiments for the WT and M213L mutated protein, respectively. (B) Plots of the decay of full-length Cx43 proteins shown in A. (C) Quantification of tau, time constant of full-length Cx43, of WT and M213L mutated protein in Triton-soluble and -insoluble fractions. *P ≤ 0.05, 2-way ANOVA, followed by Fisher’s LSD multiple-comparisons test. n = 4 and 3 independent experiments for the WT and M213L mutated protein, respectively. (D) Quantification of the ratio of pulsed full-length Cx43 in Triton-insoluble over Triton-soluble fraction at the start of the chase period (0-hour time point). **P ≤ 0.01, paired t test with 2-tailed P value. n = 4 independent experiments. (C and D) Data represent mean ± SEM.
The data in Figure 6 indicate that the M213L mutation did not decrease Cx43 protein stability. However, when comparing the τ between cytoplasmic and membrane fractions, it is striking that for both WT and mutated Cx43, Cx43 protein was 46% and 51% more stable in the Triton-insoluble fraction than in the Triton-soluble fraction (Figure 6C). These results provide corroboration that the M213L mutation did not decrease protein stability. Instead, there was more rapid degradation of poorly trafficked, cytoplasmic Cx43. Given similar mRNA and protein production rates (Figure 1C and first column of Figure 6A), the shortened half-life of cytoplasmic Cx43 resulted in less overall protein when Cx43 was not trafficked to the ICD (Figure 5D). These observations are consistent with a model where the pool of cytoplasmic Cx43 in the adult heart was degraded faster than the pool of ICD Cx43. When delivery of full-length Cx43 to the membrane was compromised due to the decrease in GJA1-20k–mediated trafficking, there was faster degradation of Cx43. In the simplest sense, Cx43 not trafficked to the ICD was degraded, resulting in decreased ICD protein and similarly decreased total Cx43 protein.
M213L mutation affects noncardiac Cx43. Cx43 is the most common of 21 human connexin proteins and has roles in physiological processes that range from reproduction and development to coordination of central nervous system activity. In our mouse models, the M213L mutation was located at the endogenous GJA1 gene. Thus, the mutated GJA1 mRNA was expressed in tissues where the GJA1 promoter was active. We quantified birth rate when breeding heterozygous animals and identified a non-Mendelian proportion of neonates: there were fewer GJA1M213L/WT and GJA1M213L/M213L pups relative to the number of GJA1WT/WT littermates (Figure 7A). We also explored Cx43 levels in mouse brains. We found, consistent with our findings in mouse hearts, that there was a similar amount of GJA1 mRNA in the brains of GJA1M213L/M213L animals (Figure 7B), yet with a reduction of Cx43 protein (Figure 7, C and D) compared with WT littermates.
 Figure 7
Figure 7The effects of M213L mutation on birth ratio and brain Cx43 expression. (A) Quantification of the number of WT, HT, and HM mice born within 6 months. The ratio is significantly different from that of the Mendelian inheritance. P ≤ 0.0001 in a χ2 test. The number of mice (n) of each genotype is indicated within bars. (B and D) The expression levels of the GJA1 mRNA (B) and Cx43 protein (D) in the brain from WT and HM mice. The data were normalized by GAPDH expression. (C) Representative Western blots showing Cx43 (FL) and GAPDH expression in the brain. All graph data represent mean ± SEM. **P ≤ 0.01 Mann-Whitney test with 2-tailed P value. n = 6 mice in each group.
-
Discussion
To explore the role of the short Cx43 isoform GJA1-20k in Cx43 trafficking, we generated a mouse model deprived of GJA1-20k using CRISPR-HDR technology to mutate the single internal methionine initiation codon for GJA1-20k (GJA1-M213L mutation). At the level of the whole animal, GJA1-20k depletion revealed highly abnormal electrocardiographic parameters, a high incidence of sudden death, and increased incidence of PVCs. At the cellular level, localization of Cx43 to ICDs was greatly reduced in the absence of GJA1-20k. We attributed the marked increase in total ventricular activation time and the decrease in QRS amplitude to a decrease in Cx43 (as the main ventricular connexin) because the level of Nav1.5 at the ICD, the other important determinant of the changes in the QRS complex, remained unchanged.
To our knowledge, the GJA1M213L/M213L mouse model we generated is the first in vivo model lacking an important Cx43 trafficking subunit (GJA1-20k) while retaining full-length Cx43 protein. Yet, there is a complementary precedent for our work. Published results involving truncated portions of the Cx43 C-terminus suggested that the C-terminus is needed to promote forward trafficking to the ICD. Cell line evidence of an independent truncated 20 kDa C-terminus band from Cx43 was first reported by the Delmar lab (8) showing that there is a significant reduction in Cx43 and a 20 kDa band in cells and mice lacking GJA1. Additionally, using a mouse containing a truncation at amino acid 258 (Cx43KO258Stop) (22) and crossing Cx43258Stop with a Cx43 knockout mouse resulted in Cx43258Stop-KO animals surviving after birth. These Cx43258Stop-KO mice had gap junction plaques that were larger, but much fewer, and overall gap junction density at the ICD was significantly decreased relative to a heterozygous Cx43-KO mouse (23). The larger size of individual plaques suggested impaired internalization due to a lack of internalization signals at the C-terminus (4, 24). Therefore, even though there was an exaggerated retention of Cx43 plaque in Cx43258Stop-KO animals, the overall number of plaques was decreased, indicating limited forward trafficking. Combining the findings of the results involving Cx43 with a truncated C-terminus with our direct evidence of the role of internally translated GJA1-20k underlines the role of the C-terminus in promoting forward trafficking to the ICD.
Our study found that the half-lives of WT Cx43 in junctional membrane fraction and nonjunctional fraction were 3.43 hours and 2.35 hours, respectively (Figure 6). Both durations are in the range of the half-life of Cx43 reported previously (4, 5, 25). However, the different half-lives indicate that the cytoplasmic pool of Cx43 was degraded approximately 50% faster than ICD Cx43. This difference in half-life between the cytoplasmic and the ICD compartments explains the overall reduced Cx43 protein level in the presence of unchanged mRNA (Figures 4 and 5) with GJA1-20k reduction. When forward trafficking was impaired, the Cx43 that did not reach the membrane was more rapidly degraded than Cx43 in the ICDs. Therapeutic interventions that rescue Cx43 forward trafficking will require time for newly synthesized Cx43 to be generated and then trafficked appropriately.
The structural integrity of gap junctions is crucial for the propagation of electrical impulses and synchronized contraction of cardiomyocytes (26, 27). However, GJA1M213L/M213L mice were able to live 2–3 weeks despite a significant reduction in the localization of Cx43 at the ICDs of ventricular myocardium (Figure 1E and Figure 3, A and B). In a prior study, mice with cardiac-specific knockout of Cx43 also started to die at 2–3 weeks, and all died within the first 2 months (28). As with this prior study (28), it is not clear why survival is possible for young mice with reduced Cx43 at their ICD. It is possible that other gap junction proteins might compensate for the lack of Cx43. Mammalian ventricular cardiomyocytes mainly express gap junction proteins Cx43 and Cx45 (29). Although the expression of Cx45 in ventricles is low under normal conditions (29), it will be interesting to examine whether its expression is upregulated in the GJA1-20k knockout mice. Cx45 is a major ventricular connexin during early development and is upregulated in heart failure (30, 31). It is also possible that, under conditions of low cell-cell Cx43 coupling, ephaptic conduction may occur (32). The GJA1M213L/M213L mouse may be a novel tool in which to study the potential of ephaptic conduction.
Cx43 has been associated with a host of noncanonical roles, from sperm motility and female reproduction (33, 34), to metabolic function (35), to neurodegenerative diseases (36). There is emerging evidence that GJA1-20k and/or other short Cx43 isoforms (instead of full-length Cx43) are involved in these physiological processes and disorders. GJA1-20k has been identified as a stress response protein that targets mitochondria (37) and mediates ischemia preconditioning (10). There is upregulation of GJA1-20k in the rat brain subject to hypoxia (12), and GJA1-20k may be involved in both the myometrium (38) and testicular function (39). Importantly, the results in Figure 7 indicate that the GJA1M213L/M213L and GJA1M213L/WT mice may represent useful models to further explore the role of GJA1-20k in well-recognized but poorly understood noncanonical functions of Cx43.
Our data indicated that the M213L mutation did not significantly change the half-life of the full-length Cx43 protein in either the junctional or nonjunctional fraction (Figure 6). It also has been identified in vitro that the presence or absence of GJA1-20k determines whether Cx43 arrives at cell-cell borders, whether WT Cx43 or GJA1-M213L plasmids are used (6). However, it is possible that the M213L mutation affects other biological functions of Cx43, in addition to limiting the translation of GJA1-20k. The GJA1M213L mutant mice, and cells from this mouse, will be a valuable tool to test this possibility.
Taken together, we have developed a GJA1M213L/M213L mouse model that effectively removed endogenous GJA1-20k (while preserving full-length Cx43) by mutating a single AUG internal ribosomal start site. We have found that the GJA1M213L/M213L mice experienced severe defects in cardiac electrical coupling, reductions in total Cx43 protein, and sudden death as a result of impaired delivery of full-length Cx43 to the ICDs. The significantly shorter lifespan of cytosolic Cx43 versus Cx43 at the ICD explains why Cx43 connexons that were not trafficked to the ICD were rapidly degraded and led to a decrease of overall cellular Cx43 protein.
Emerging data have underscored the stress-related upregulation of GJA1-20k and its modulatory effects on ion channel trafficking (6, 7), cytoskeleton organization (7), metabolic regulation (10, 37), and cardioprotection (10). The GJA1M213L/M213L mouse model will be useful to explore additional physiological functions of this internally translated protein.
-
Methods
Generation of the GJA1-M213L knockin founders and genotyping. CRISPR target sequence around the targeted mutation site was selected using the CRISPOR web algorithm (http://crispor.tefor.net) (40). We chose a CRISPR target sequence of ATTCAGAGCGAGAGACACGA followed by a PAM (AGG), of which the potential cleavage site is located at 16 bases downstream to the targeted codon (ATG to TTA; M213L). The crRNA (AUUCAGAGCGAGAGACACCAGUUUUAGAGCUAUGCUGUUUUG) and tracrRNA (U-002000-120) were synthesized by Dharmacon, Inc. A donor oligo that contained the desired mutation with 60 bases homology arms in both sides that was complementary to the CRISPR target was synthesized by Integrated DNA Technologies, Inc. We included a point mutation (TCC to TCG) for disruption of the PAM to avoid reediting after successful induction of HDR.
A CRISPR mixture consisting of 25 ng/μL oligo donor, 60 ng/μL crRNA/tracrRNA mix (1:1 molar ratio), and 50 ng/μL eSpCas9 protein (ESPCAS9PRO-50UG) (MilliporeSigma) was introduced into C57BL/6J (Strain 000664, The Jackson Laboratory) fertilized eggs by pronuclear microinjection with a standard method (41). Validation of efficacy of the guide RNA and HDR was performed by PCR genotyping on blastocysts in the initial phase and subsequently on tissue samples isolated from transgenic founders and progeny. Briefly, single blastocysts or toe/tail tissue samples were processed and amplified using the KAPA genotyping kit (KAPA Bioscience, KK7352) with PCR primers (TGGGATTGAAGAACACGGCA and CCACGATAGCTAAGGGCTGG) flanking the targeted mutation site. A 668 bp PCR fragment was isolated from a preparative gel and purified with QIAquick PCR purification kit (QIAGEN) for digestion with NlaIII restriction enzyme. WT band segregates into 227 bp and 441 bp fragments because of the native NlaIII site around the targeted site, while the mutant band remains at 668 bp as this site is destroyed upon successful introduction of desired or undesired mutation(s). Status of introduced mutation(s) was further confirmed by sequencing of the PCR fragments.
We confirmed 9 recombinant mice with heterozygous or mosaic mutations out of 21 mice produced. We established 2 lines (founder lines 37 and 51) after 2 generations of backcrossing with WT C57BL/6J mice to dilute out possible off-targeting mutations and mosaicism. The 2 lines were used for further expansion of colonies and experiments.
The oligo donor with mutations (underlined) is as follows: CCCCACCAGGTGGACTGCTTCCTCTCACGTCCCACGGAGAAAACCATCTTCATCATCTTCTTACTGGTGGTGTCGTTGGTGTCTCTCGCTCTGAATATCATTGAGCTCTTCTATGTCTTCTTC.
Quantitative RT-PCR. Total RNA was extracted from cardiac ventricular or brain tissues of 2- to 3-week-old mice using Trizol per the manufacturer’s instructions (Thermo Fisher Scientific, 15596018), followed by an additional purification step with the PureLink RNA mini kit (Thermo Fisher Scientific, 12183018A) and DNase treatment (TURBO DNase, Thermo Fisher Scientific, AM1907). Purified total RNA was reverse transcribed into cDNA with the SuperScript IV VILO master mix (Thermo Fisher Scientific, 11756050). TaqMan probes (Thermo Fisher Scientific) to the mouse GJA1 (assay ID: Mm00439105-m1) and GAPDH (assay ID: Mm99999915-g1) genes were used to amplify and detect the corresponding genes. The thermocycling setup was 2 minutes at 50°C; 10 minutes at 95°C; 40 cycles of 15 seconds at 95°C; and 1 minute at 60°C. Relative expression of the GJA1 mRNA in mice with different genotypes was normalized to the GAPDH mRNA expression level and quantified using the ΔCt method.
Immunoprecipitation of cardiac Cx43 isoforms. Two- to 3-week-old mouse hearts were isolated and perfused with HBSS. Ventricles were homogenized and proteins were extracted in lysis buffer (50 mM Tris pH 8, 5 mM EDTA, 150 mM NaCl, 10 mM KCl, 0.75% Triton X-100, 5 mM NaF, 1 mM NaVO3) supplemented with Halt protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, 78445) at 4°C for 2 hours (42). Homogenate was centrifuged at 16,000 g at 4°C for 25 minutes. Lysate supernatant (equivalent to 1.5 mg of protein) was diluted 2-fold with lysis buffer without triton and then precleared with Dynabeads Protein G (Thermo Fisher Scientific, 10004D) at 4°C for 30 minutes. Precleared lysate was incubated with 5 μg of mouse anti-Cx43CT1 antibody (Dr. Paul Lampe, the Fred Hutchinson Cancer Research Center) at 4°C overnight. Samples were subsequently incubated with Dynabeads Protein G at 4°C for an hour. Immunoprecipitates were washed with buffer (50 mM Tris pH 8, 5 mM EDTA, 150 mM NaCl, 10 mM KCl, 0.25% Triton X-100, 5 mM NaF, 1 mM NaVO3) supplemented with Halt protease and phosphatase inhibitor cocktail, then eluted in 2x sample buffer. The eluates were subjected to Western blotting.
Western blotting. Western blotting was performed as described in Basheer et al. (7). Protein samples were subjected to SDS-PAGE electrophoresis using NuPAGE Bis-Tris gels (Thermo Fisher Scientific, NP0335BOX) in MES-SDS running buffer (Thermo Fisher Scientific, NP0002). NuPAGE Tris-Acetate gels (Thermo Fisher Scientific, EA0375BOX) and Tris-Acetate running buffer (Thermo Fisher Scientific, LA0041) were used for detection of high molecular weight protein such as Nav1.5. The gels were electroblotted onto FluoroTrans PVDF membranes (Pall Corporation, BSP0161). For Western blotting with antibodies, membranes were blocked at room temperature with 5% nonfat dry milk (Carnation) in TNT buffer (50 mM Tris pH 8.0, 150 mM NaCl, 0.1% Tween-20), probed at 4°C overnight with primary antibodies in blocking buffer, and incubated with fluorophore-labeled secondary antibody in blocking buffer at room temperature for an hour. For Western blotting with streptavidin, membranes were blocked at room temperature with 5% BSA (MilliporeSigma, A9647) in TNT buffer, and probed with Alexa Fluor 555 streptavidin (1 to 500 dilution, Thermo Fisher Scientific, S32355) in blocking buffer at 4°C overnight.
Western blots were visualized using the ChemiDoc MP System and quantified with Image Lab software (Bio-Rad) or ImageJ. Primary antibodies used were chicken anti-Cx43 antibody (custom made, detailed below), rabbit anti-Cx43 (1 to 2000 dilution, MilliporeSigma, C6219), mouse anti-Cx43 antibody (1 to 1000 dilution, Millipore, MABT902, clone P2C4), mouse anti–N-cadherin (1 to 1000 dilution, BD Biosciences, 610921), rat anti–α-tubulin (1 to 2000 dilution, Abcam, ab6160), rabbit anti-Nav1.5 (1 to 200 dilution, provided by Dr. Hugues Abriel at the University of Bern, Bern, Switzerland), and mouse anti-GAPDH (1 to 2000 dilution, Abcam, ab8245).
Generation of chicken anti-Cx43 antibody. A peptide, cdqrpssrassrassrprpddlei, which corresponds to the amino acid sequence 360-382 of the human Cx43 protein and an extra cysteine at the N-terminus, was synthesized (Bio-Synthesis, Texas) and used as the antigen to produce mouse monoclonal antibodies (Epitope Recognition and Immunoreagent Core Facility, University of Alabama at Birmingham, Birmingham, Alabama, USA). The variable regions of the heavy and light chains of a mouse monoclonal antibody were sequenced and fused with the chicken IgY and chicken lambda-like constant regions, respectively, to generate the chicken anti-Cx43 antibody (Oak Biosciences).
ECG recording and analysis. Two- to 3-week-old mice were anesthetized with isoflurane and positioned in dorsal recumbency position. Electrodes were attached to the left and right forelimbs and left hind limb in a bipolar lead 1 configuration. ECGs were recorded with the ADInstruments PowerLab hardware (ADInstruments) for 5 minutes after 5 minutes of stabilization of the system. Data were analyzed using the ECG analysis module in LabChart8 software (ADInstruments). The R wave amplitude and QRS duration of each trace were the average values of all the beats in 3 to 5 minutes of recording. In a case where the software could not detect the correct QRS complexes of a homozygous GJA1-20k knockout mouse, the R wave amplitudes and QRS duration of 40 beats or greater were manually measured and averaged.
Telemetry recording and analysis. Telemetry recording was performed using the DSI MX2 telemetry acquisition system (Data Sciences International) and LabChart8 software (ADInstruments) according to the manufacturer’s instructions. Briefly, 6- to 7-month-old adult mice were anesthetized with isoflurane and positioned in dorsal recumbency. A telemetry implant (DSI PhysioTel ETA-F10 for Mice, Data Sciences International) was surgically placed subcutaneously along the lateral flank, with leads oriented cranially. The ends of the negative and positive leads were tunneled subcutaneously to the right pectoral muscle and left caudal rib region, respectively. Both leads were secured to the abdominal wall with a stay suture. The skin incision was closed with suture. After the mice returned to normal postures and behaviors, their ECG, activities, and temperature were recorded overnight, following their regular light/dark cycles.
Continuous recordings of 3 hours were started between 9 pm and 11 pm in all mice to exclude influences of diurnal rhythms. Data in ASCI format were analyzed with custom written software using LabVIEW (National Instruments Inc.) and MATLAB (The Mathworks, Inc.). Importantly, all ECG complexes were analyzed continuously, first by an automatic algorithm derived from Holter analysis software (Schiller Inc.), and subsequently by observing the totality of electrical heartbeats on a graphical user interface. Overall, a single data recording of 3 hours included approximately 108,000 electrical heartbeats.
Echocardiography and analysis. Transthoracic echocardiography imaging was performed on anesthetized animals using a VEVO 3100 ultrasound machine with MX5550D transducer (VisualSonics) (10). LV volume and percentage of ejection fraction (EF) and fractional shortening (FS) were determined from the B-mode long axis view. LVID (left ventricular internal diameter), LVPW (left ventricular posterior wall thickness), LVAW (left ventricular anterior wall thickness), heart rate, and LV mass measurements were determined from the M-mode short axis view. E/A and E/e’ ratios were measured by Doppler assessment.
Immunostaining, confocal microscopy, and quantification of Cx43 at the ICDs. Cryosections (5 μm) of cardiac ventricular tissues were fixed in prechilled acetone for 10 minutes on ice, blocked, and permeabilized at room temperature for an hour in 10% normal goat serum, 2% IgG-free BSA, 0.2% Triton X-100 in 1x PBS. The sections were incubated with primary antibodies in antibody diluent (10% normal goat serum, 0.1% Triton X-100 in 1x PBS) at 4°C overnight. The sections were then incubated with fluorophore conjugated secondary antibodies (1:500, Thermo Fisher Scientific) and mounted using Fluoromount-G mounting medium (Southern Biotech, 0100-01). The primary antibodies used were rabbit anti-Cx43 (N-term) (1:100, OriGene, AP11568PU-N), rabbit anti-Nav1.5 (1:50, kindly gifted from Dr. Hugues Abriel at the University of Bern, Bern, Switzerland), and mouse anti–N-Cadherin (1:100, BD Biosciences, 610921).
The immunostained tissue sections were imaged using a Nikon Eclipse Ti imaging system with a ×60/1.49 Apo objective. Z-stacks (5 μm of total length) of confocal images were obtained.
Quantification of Cx43 or Nav1.5 expression at the ICDs, which were marked by N-cadherin staining, was performed as previously described (7). Average intensity projection of each stack of confocal images was processed in ImageJ and used to assess protein expression. N-cadherin images were used to generate binary masks of ICDs, which were subsequently image-multiplied by the corresponding Cx43 image. Cx43 or Nav1.5 fluorescence signal at the ICDs was measured and compared among mice of different genotypes.
Preparation of cardiac ventricular and brain lysates. Mouse cardiac ventricular or brain tissues were homogenized and lysed in RIPA buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 1 mM NaF, 0.2 mM Na3VO4) supplemented with Halt protease and phosphatase inhibitor cocktail. Samples were incubated at 4°C for 2 hours, followed by centrifugation at 16,000 g at 4°C for 25 minutes. The lysate supernatant was collected, quantified, and subjected to Western blotting.
Triton solubility fractionation (in vivo). Triton fractionation was performed as previously described (18). Briefly, the snap-frozen heart tissues from 2-week-old mice were homogenized and rotated at 4°C for 1 hour in 1% Triton X-100 buffer (50 mM Tris pH 7.4, 1% Triton X-100, 2 mM EDTA, 2 mM EGTA, 250 mM NaCl, 1 mM NaF, 0.1 mM Na3VO4) with Halt protease and phosphatase inhibitor cocktail at a final concentration of 100 mg tissue/mL. The lysates were centrifuged for 20 minutes at 10,000 g in preweighed tubes. Supernatant was collected and added to an equal volume of 1% Triton X-100 buffer containing 8 M urea and 2 M thiourea for the soluble fraction. The pellets were weighed and suspended in 1% Triton X-100 buffer containing 4 M urea and 1 M thiourea at a final concentration of 30 mg/mL. NuPAGE sample buffer was added to all fractions followed by sonication. DTT at a final concentration of 100 mM was added and incubated at 37 °C for 20 minutes to reduce the samples, followed by centrifuge at 10,000 g for 20 minutes. The supernatant from soluble or insoluble fraction was subjected to Western blotting described above.
Cloning of mouse Cx43 constructs. WT and M213L mutated mouse GJA1 DNA were amplified from the total cardiac cDNA of a WT and a homozygous GJA1-20k knockout mouse, respectively, using primers that contained sequences resistant to the Cx43 siRNA. The amplicons were cloned into pDONR 221 (Thermo Fisher Scientific, 12536017) and pcDNA3.2/HA-DEST (9) using Gateway cloning (Thermo Fisher Scientific) to generate C-terminally HA-tagged constructs.
Primers used in PCR amplification. The primers were as follows: Cx43 Fwd 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGCCGCCACCATGGGTGACTGGAGCGCC-3′; Mouse Cx43 CDS Resist Fwd 5′-ATGGGTGACTGGAGCGCCTTGGGGAAGCTGCTGGACAAGGTCCAAGCCTACTCCACGGCCGGAGGGAAGGTGTGGCTGTCGGTGCTCTTCATTTTCAGAATCCTGCTCCTGGGGACAGCGGTTGAGTCAGCTTGGGGTGACGAACAATCCGCATTCCGCTGTAACACTCAACAACCCG-3′; Mouse Cx43 Rev 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTAAATCTCCAGGTCATCAGGCCG-3′.
Pulse-chase assay with Click-iT chemistry and Triton fractionation. HEK293FT cells (Thermo Fisher Scientific) were maintained in DMEM supplemented with 10% FBS, nonessential amino acids, sodium pyruvate (Thermo Fisher Scientific), and Mycozap-CL (Lonza, VZA-2012) unless specified.
Pulse-chase assay. The pulse-chase experiment was adapted from the procedure described by James et al. (5) with the following modifications. Hek293T cells were reverse transfected with human GJA1 siRNA (Thermo Fisher Scientific, Assay ID: HSS178257) (6) at a final concentration of 16 nM using RNAiMax (Thermo Fisher Scientific, 13778150) and plated in 10-cm dishes. Two days after siRNA transfection, siRNA-resistant mouse Cx43 constructs were transfected using FuGENE HD Transfection Reagent (Promega, E2312). One day after plasmid transfection, cells were washed and starved for 1 hour in methionine/cysteine-free DMEM (Thermo Fisher Scientific, 21013024) supplemented with 10% dialyzed FBS (GE Healthcare, SH30079.01 or Thermo Fisher Scientific, A3382001). Cells were pulsed for 1 hour with Click-iT L-Azidohomoalanine (Thermo Fisher Scientific, C10102) at a final concentration of 50 μM added in starvation media. Cells were returned to normal growth medium with extra methionine (1 mM, MilliporeSigma, M5308) and cysteine (1 mM, MilliporeSigma, C7352). Cells were harvested over a 7.5-hour period at 2.5-hour intervals by centrifuging at 200 g for 5 minutes.
Triton solubility fractionation (in vitro). Cell pellet from a 10-cm dish was solubilized in 400 μL of Triton lysis buffer (50 mM Tris, pH 7.4, 1% Triton X-100, 2 mM EDTA, 2 mM EGTA, 250 mM NaCl) supplemented with Halt protease and phosphatase inhibitor cocktail at 4°C for an hour. An aliquot of lysate (60 μL) was saved as total protein fraction. The rest of the lysate was centrifuged at 15,000 g for 30 minutes. The supernatant and pellet served as the Triton-soluble and -insoluble fractions, respectively.
The total protein fraction was mixed with 4x LDS sample buffer (Thermo Fisher Scientific, NP0007) with DTT, incubated at 37°C for 20 minutes, briefly sonicated, and centrifuged at 10,000 g for 20 minutes. The supernatant was subjected to Western blotting.
Immunoprecipitation and labeling of pulsed proteins using Click-iT chemistry. The Triton-soluble fraction (~340 μL) was sonicated and diluted with 227 μL of Triton lysis buffer without NaCl to reduce salt concentration. The lysate was precleared with Dynabeads Protein G at 4°C for 30 minutes and incubated with 3 μg of mouse anti-HA antibody (Abcam, ab1424, clone 4C12) or 2 μg of rabbit anti-Cx43 antibody (MilliporeSigma, C6219) at 4°C overnight. Samples were then incubated with Dynabeads Protein G at 4°C for an hour. Immunoprecipitates were washed 4 times for 10 minutes with wash buffer (50 mM Tris, pH 7.4, 1% Triton X-100, 2 mM EDTA, 2 mM EGTA, 150 mM NaCl) supplemented with Halt protease and phosphatase inhibitor cocktail. Each immunoprecipitate was washed once and resuspended in 30 μL of 50 mM Tris pH 8. The pulsed proteins in the immunoprecipitate were labeled with biotin-alkyne using the Click-iT Biotin Protein Analysis Detection Kit (Thermo Fisher Scientific, C33372) in a final volume of 100 μL according to the manufacturer’s recipe at 4°C for an hour (Thermo Fisher Scientific, ref. 43). The labeled immunoprecipitate was washed twice in TBS (50 mM Tris-HCl, 150 mM NaCl, pH 7.5). The bound proteins were eluted in 30 μL of 2x LDS buffer with DTT at 37°C for 20 minutes and subjected to Western blotting. Pulsed and biotin-labeled proteins were detected with Alexa Fluor 555 streptavidin (Thermo Fisher Scientific, S32355). Total Cx43 in the immunoprecipitates was also probed for with rabbit anti-Cx43 antibody (MilliporeSigma, C6219) or mouse anti-Cx43 antibody (Millipore, MABT902, clone P2C4).
The Triton-insoluble fraction was redissolved in 50 μL of 50 mM Tris-HCl pH 7.4, 2% SDS, 1 mM EDTA supplemented with Halt protease and phosphatase inhibitor cocktail at 37°C for 10 minutes with pipetting. The solubilized fraction was diluted with 7 volume of 150 mM NaCl, 50 mM Tris-HCl pH 7.4, 1 mM EDTA, 0.6% NP-40 supplemented with Halt protease and phosphatase inhibitor cocktail. The solution was sonicated and centrifuged at 10,000 g at 4°C for 20 minutes. The supernatant was immunoprecipitated and labeled with Click-iT biotin-alkyne as described above for the Triton-soluble fraction. The bound proteins were eluted and subjected to Western blotting as described above.
Partition of newly synthesized Cx43 in Triton-soluble and -insoluble fractions. The amount of pulsed Cx43, detected by streptavidin on a Western blot, at the 0-hour time point was normalized to the amount of total Cx43 in the corresponding immunoprecipitate to account for the difference in immunoprecipitation efficiency. The normalized pulsed Cx43 in the Triton-insoluble fraction was divided by that in the Triton-soluble fraction, which served as an indication of the ratio between junctional and nonjunctional Cx43. This ratio in cells transfected with the M213L mutated Cx43 was compared with that of the WT Cx43 to assess the effect of the mutation on delivery of Cx43 to cell-cell junctions.
Quantification of the time constant of full-length Cx43. At different time points during the chase period, the normalized pulsed Cx43 (calculated as described above) was divided by that at the 0-hour time point in a specified fraction. Percentages of the remaining normalized pulsed Cx43 were fit with an exponential line in Microsoft Excel. The time constant (τ) was deduced from the exponential equation, y = ae–t/τ. Half-lives were calculated with the equation, half-life = τ*ln2.
Statistics. All quantitative data were expressed as mean ± SEM. The 2-tailed Student’s t test, 1-way ANOVA with multiple-comparisons test, 2-way ANOVA with multiple-comparisons test, Kaplan-Meier survival curves, and comparisons were performed as appropriate using Prism 6 software (GraphPad). A P value less than 0.05 was considered significant.
Study approval. All mice were maintained under sterile barrier conditions. All procedures were reviewed and approved by Cedars-Sinai Medical Center and the University of Utah Institutional Animal Care and Use Committee.
-
Author contributions
SX, DS, AGK, TTH, and RMS designed research studies. SX, DS, RB, and DMH conducted experiments, acquired data, and analyzed data. SA conducted experiments. YK and MK designed and conducted experiments related to production of knockout mice. PDL provided reagents. AGK analyzed data. SX, DS, and RMS wrote the manuscript. SX, DS, RB, DMH, SA, YK, MK, PDL, AGK, TTH, and RMS reviewed and edited the manuscript. AGK, TTH, and RMS supervised the study.
-
Acknowledgments
We thank TC Hitzeman for advice on statistical analysis. Hugues Abriel at the University of Bern provided anti-Nav1.5 antibody. This work was supported by NIH/NHLBI grants (AGK-HL136463, TTH-HL133286, RMS-HL138577, and RMS-HL152691).
Address correspondence to: Robin M. Shaw, Nora Eccles Harrison Cardiovascular Research and Training Institute, University of Utah, 95 South 2000 East, Suite 143, Salt Lake City, Utah 84112, USA. Phone: 801.581.8183; Email: Robin.Shaw@hsc.utah.edu.
-
Footnotes
TTH’s present address is: Department of Pharmacology & Toxicology, College of Pharmacy, University of Utah, Salt Lake City, Utah, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(9):4858–4870.https://doi.org/10.1172/JCI134682.
-
References
- Basheer W, Shaw R. The “tail” of connexin43: an unexpected journey from alternative translation to trafficking. Biochim Biophys Acta. 2016;1863(7 pt B):1848–1856.View this article via: PubMed Google Scholar
- Lo CW. Role of gap junctions in cardiac conduction and development: insights from the connexin knockout mice. Circ Res. 2000;87(5):346–348.
- Saffitz JE. Arrhythmogenic cardiomyopathy and abnormalities of cell-to-cell coupling. Heart Rhythm. 2009;6(8 suppl):S62–S65.View this article via: PubMed Google Scholar
- Smyth JW, et al. A 14-3-3 mode-1 binding motif initiates gap junction internalization during acute cardiac ischemia. Traffic. 2014;15(6):684–699.
- James CC, Zeitz MJ, Calhoun PJ, Lamouille S, Smyth JW. Altered translation initiation of Gja1 limits gap junction formation during epithelial-mesenchymal transition. Mol Biol Cell. 2018;29(7):797–808.
- Smyth JW, Shaw RM. Autoregulation of connexin43 gap junction formation by internally translated isoforms. Cell Rep. 2013;5(3):611–618.
- Basheer WA, et al. GJA1-20k arranges actin to guide Cx43 delivery to cardiac intercalated discs. Circ Res. 2017;121(9):1069–1080.
- Joshi-Mukherjee R, Coombs W, Burrer C, de Mora IA, Delmar M, Taffet SM. Evidence for the presence of a free C-terminal fragment of cx43 in cultured cells. Cell Commun Adhes. 2007;14(2–3):75–84.View this article via: PubMed Google Scholar
- Smyth JW, et al. Actin cytoskeleton rest stops regulate anterograde traffic of connexin 43 vesicles to the plasma membrane. Circ Res. 2012;110(7):978–989.
- Basheer WA, et al. Stress response protein GJA1-20k promotes mitochondrial biogenesis, metabolic quiescence, and cardioprotection against ischemia/reperfusion injury. JCI Insight. 2018;3(20):121900.
- Salat-Canela C, Sesé M, Peula C, Ramón y Cajal S, Aasen T. Internal translation of the connexin 43 transcript. Cell Commun Signal. 2014;12:31. View this article via: PubMed Google Scholar
- Ul-Hussain M, et al. Internal ribosomal entry site (IRES) activity generates endogenous carboxyl-terminal domains of Cx43 and is responsive to hypoxic conditions. J Biol Chem. 2014;289(30):20979–20990.
- Reaume AG, et al. Cardiac malformation in neonatal mice lacking connexin43. Science. 1995;267(5205):1831–1834.
- Bacharova L, Mateasik A, Krause R, Prinzen FW, Auricchio A, Potse M. The effect of reduced intercellular coupling on electrocardiographic signs of left ventricular hypertrophy. J Electrocardiol. 2011;44(5):571–576.
- Danik SB, et al. Modulation of cardiac gap junction expression and arrhythmic susceptibility. Circ Res. 2004;95(10):1035–1041.
- Morley GE, et al. Reduced intercellular coupling leads to paradoxical propagation across the Purkinje-ventricular junction and aberrant myocardial activation. Proc Natl Acad Sci U S A. 2005;102(11):4126–4129.
- Bacharova L, Plandorova J, Klimas J, Krenek P, Kyselovic J. Discrepancy between increased left ventricular mass and “normal” QRS voltage is associated with decreased connexin 43 expression in early stage of left ventricular hypertrophy in spontaneously hypertensive rats. J Electrocardiol. 2008;41(6):730–734.
- Smyth JW, et al. Limited forward trafficking of connexin 43 reduces cell-cell coupling in stressed human and mouse myocardium. J Clin Invest. 2010;120(1):266–279.
- Asimaki A, et al. A new diagnostic test for arrhythmogenic right ventricular cardiomyopathy. N Engl J Med. 2009;360(11):1075–1084.
- Shaw RM, Fay AJ, Puthenveedu MA, von Zastrow M, Jan YN, Jan LY. Microtubule plus-end-tracking proteins target gap junctions directly from the cell interior to adherens junctions. Cell. 2007;128(3):547–560.
- Shaw RM, Rudy Y. Ionic mechanisms of propagation in cardiac tissue. Roles of the sodium and L-type calcium currents during reduced excitability and decreased gap junction coupling. Circ Res. 1997;81(5):727–741.
- Maass K, et al. Defective epidermal barrier in neonatal mice lacking the C-terminal region of connexin43. Mol Biol Cell. 2004;15(10):4597–4608.
- Maass K, Shibayama J, Chase SE, Willecke K, Delmar M. C-terminal truncation of connexin43 changes number, size, and localization of cardiac gap junction plaques. Circ Res. 2007;101(12):1283–1291.
- Remo BF, et al. Phosphatase-resistant gap junctions inhibit pathological remodeling and prevent arrhythmias. Circ Res. 2011;108(12):1459–1466.
- Beardslee MA, Laing JG, Beyer EC, Saffitz JE. Rapid turnover of connexin43 in the adult rat heart. Circ Res. 1998;83(6):629–635.
- Delorme B, et al. Expression pattern of connexin gene products at the early developmental stages of the mouse cardiovascular system. Circ Res. 1997;81(3):423–437.
- Kasi VS, et al. Cardiac-restricted angiotensin-converting enzyme overexpression causes conduction defects and connexin dysregulation. Am J Physiol Heart Circ Physiol. 2007;293(1):H182–H192.
- Gutstein DE, et al. Conduction slowing and sudden arrhythmic death in mice with cardiac-restricted inactivation of connexin43. Circ Res. 2001;88(3):333–339.
- van Veen AA, van Rijen HV, Opthof T. Cardiac gap junction channels: modulation of expression and channel properties. Cardiovasc Res. 2001;51(2):217–229.
- SVaidya D, et al. Null mutation of connexin43 causes slow propagation of ventricular activation in the late stages of mouse embryonic development. Circ Res. 2001;88(11):1196–1202.
- Yamada KA, Rogers JG, Sundset R, Steinberg TH, Saffitz J. Up-regulation of connexin45 in heart failure. J Cardiovasc Electrophysiol. 2003;14(11):1205–1212.
- Veeraraghavan R, Gourdie RG, Poelzing S. Mechanisms of cardiac conduction: a history of revisions. Am J Physiol Heart Circ Physiol. 2014;306(5):H619–H627.
- Kidder GM, Cyr DG. Roles of connexins in testis development and spermatogenesis. Semin Cell Dev Biol. 2016;50:22–30.
- Winterhager E, Kidder GM. Gap junction connexins in female reproductive organs: implications for women’s reproductive health. Hum Reprod Update. 2015;21(3):340–352.
- Rodríguez-Sinovas A, Ruiz-Meana M, Denuc A, García-Dorado D. Mitochondrial Cx43, an important component of cardiac preconditioning. Biochim Biophys Acta Biomembr. 2018;1860(1):174–181.
- Freitas-Andrade M, Naus CC. Astrocytes in neuroprotection and neurodegeneration: the role of connexin43 and pannexin1. Neuroscience. 2016;323:207–221.
- Fu Y, et al. Cx43 isoform GJA1-20k promotes microtubule dependent mitochondrial transport. Front Physiol. 2017;8:905. View this article via: PubMed Google Scholar
- Nadeem L, Shynlova O, Mesiano S, Lye S. Progesterone via its type-A receptor promotes myometrial gap junction coupling. Sci Rep. 2017;7(1):13357.
- Kubincová P, et al. Polycyclic aromatic hydrocarbons and endocrine disruption: role of testicular gap junctional intercellular communication and connexins. Toxicol Sci. 2019;169(1):70–83.
- Haeussler M, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17(1):148.
- Behringer R, Gertsenstein M, Nagy KV, Nagy A. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor Laboratory Press; 2014.
- Fu Y, et al. Isoproterenol promotes rapid ryanodine receptor movement to bridging integrator 1 (BIN1)-organized dyads. Circulation. 2016;133(4):388–397.
- Antinone SE, Ghadge GD, Lam TT, Wang L, Roos RP, Green WN. Palmitoylation of superoxide dismutase 1 (SOD1) is increased for familial amyotrophic lateral sclerosis-linked SOD1 mutants. J Biol Chem. 2013;288(30):21606–21617.
- Basheer W, Shaw R. The “tail” of connexin43: an unexpected journey from alternative translation to trafficking. Biochim Biophys Acta. 2016;1863(7 pt B):1848–1856.
-
Version history
- Version 1 (June 11, 2020): In-Press Preview
- Version 2 (August 10, 2020): Electronic publication
- Version 3 (September 1, 2020): Print issue publication



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)