Advertisement
Research ArticleGeneticsOncology Free access | 10.1172/JCI129941
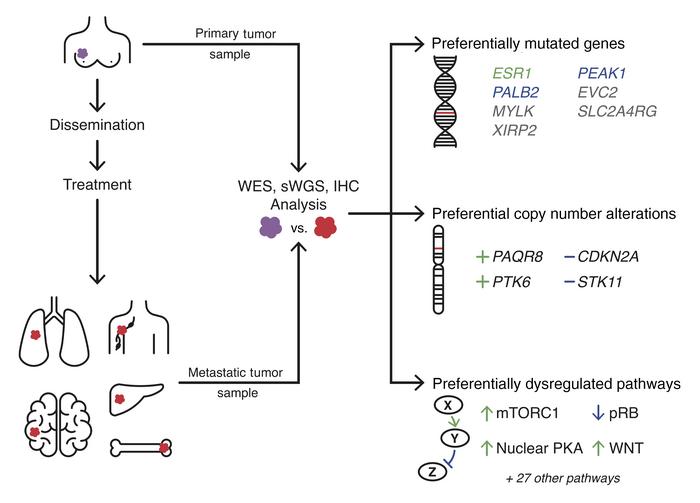
Genomic landscape of metastatic breast cancer identifies preferentially dysregulated pathways and targets
Matt R. Paul,1,2,3 Tien-chi Pan,1,2,3 Dhruv K. Pant,1,2,3 Natalie N.C. Shih,1,4 Yan Chen,1,2,3 Kyra L. Harvey,1,2,3 Aaron Solomon,1,2,3 David Lieberman,4 Jennifer J.D. Morrissette,4 Danielle Soucier-Ernst,1,5 Noah G. Goodman,1,5 S. William Stavropoulos,1,6 Kara N. Maxwell,1,5 Candace Clark,1,5 George K. Belka,1,2,3 Michael Feldman,1,4 Angela DeMichele,1,5,7 and Lewis A. Chodosh1,2,3,5
1Secondary Prevention through Surveillance and Intervention (2-PREVENT) Translational Center of Excellence,
2Abramson Family Cancer Research Institute,
3Department of Cancer Biology,
4Department of Pathology and Laboratory Medicine,
5Department of Medicine,
6Department of Radiology, and
7Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Address correspondence to: Lewis A. Chodosh, Room 614 BRB II/III, 421 Curie Boulevard, Philadelphia, Pennsylvania 19104-6160, USA. Email: chodosh@pennmedicine.upenn.edu.
Find articles by Paul, M. in: PubMed | Google Scholar
1Secondary Prevention through Surveillance and Intervention (2-PREVENT) Translational Center of Excellence,
2Abramson Family Cancer Research Institute,
3Department of Cancer Biology,
4Department of Pathology and Laboratory Medicine,
5Department of Medicine,
6Department of Radiology, and
7Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Address correspondence to: Lewis A. Chodosh, Room 614 BRB II/III, 421 Curie Boulevard, Philadelphia, Pennsylvania 19104-6160, USA. Email: chodosh@pennmedicine.upenn.edu.
Find articles by
Pan, T.
in:
PubMed
|
Google Scholar
|

1Secondary Prevention through Surveillance and Intervention (2-PREVENT) Translational Center of Excellence,
2Abramson Family Cancer Research Institute,
3Department of Cancer Biology,
4Department of Pathology and Laboratory Medicine,
5Department of Medicine,
6Department of Radiology, and
7Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Address correspondence to: Lewis A. Chodosh, Room 614 BRB II/III, 421 Curie Boulevard, Philadelphia, Pennsylvania 19104-6160, USA. Email: chodosh@pennmedicine.upenn.edu.
Find articles by Pant, D. in: PubMed | Google Scholar
1Secondary Prevention through Surveillance and Intervention (2-PREVENT) Translational Center of Excellence,
2Abramson Family Cancer Research Institute,
3Department of Cancer Biology,
4Department of Pathology and Laboratory Medicine,
5Department of Medicine,
6Department of Radiology, and
7Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Address correspondence to: Lewis A. Chodosh, Room 614 BRB II/III, 421 Curie Boulevard, Philadelphia, Pennsylvania 19104-6160, USA. Email: chodosh@pennmedicine.upenn.edu.
Find articles by Shih, N. in: PubMed | Google Scholar
1Secondary Prevention through Surveillance and Intervention (2-PREVENT) Translational Center of Excellence,
2Abramson Family Cancer Research Institute,
3Department of Cancer Biology,
4Department of Pathology and Laboratory Medicine,
5Department of Medicine,
6Department of Radiology, and
7Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Address correspondence to: Lewis A. Chodosh, Room 614 BRB II/III, 421 Curie Boulevard, Philadelphia, Pennsylvania 19104-6160, USA. Email: chodosh@pennmedicine.upenn.edu.
Find articles by Chen, Y. in: PubMed | Google Scholar
1Secondary Prevention through Surveillance and Intervention (2-PREVENT) Translational Center of Excellence,
2Abramson Family Cancer Research Institute,
3Department of Cancer Biology,
4Department of Pathology and Laboratory Medicine,
5Department of Medicine,
6Department of Radiology, and
7Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Address correspondence to: Lewis A. Chodosh, Room 614 BRB II/III, 421 Curie Boulevard, Philadelphia, Pennsylvania 19104-6160, USA. Email: chodosh@pennmedicine.upenn.edu.
Find articles by Harvey, K. in: PubMed | Google Scholar
1Secondary Prevention through Surveillance and Intervention (2-PREVENT) Translational Center of Excellence,
2Abramson Family Cancer Research Institute,
3Department of Cancer Biology,
4Department of Pathology and Laboratory Medicine,
5Department of Medicine,
6Department of Radiology, and
7Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Address correspondence to: Lewis A. Chodosh, Room 614 BRB II/III, 421 Curie Boulevard, Philadelphia, Pennsylvania 19104-6160, USA. Email: chodosh@pennmedicine.upenn.edu.
Find articles by Solomon, A. in: PubMed | Google Scholar
1Secondary Prevention through Surveillance and Intervention (2-PREVENT) Translational Center of Excellence,
2Abramson Family Cancer Research Institute,
3Department of Cancer Biology,
4Department of Pathology and Laboratory Medicine,
5Department of Medicine,
6Department of Radiology, and
7Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Address correspondence to: Lewis A. Chodosh, Room 614 BRB II/III, 421 Curie Boulevard, Philadelphia, Pennsylvania 19104-6160, USA. Email: chodosh@pennmedicine.upenn.edu.
Find articles by Lieberman, D. in: PubMed | Google Scholar
1Secondary Prevention through Surveillance and Intervention (2-PREVENT) Translational Center of Excellence,
2Abramson Family Cancer Research Institute,
3Department of Cancer Biology,
4Department of Pathology and Laboratory Medicine,
5Department of Medicine,
6Department of Radiology, and
7Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Address correspondence to: Lewis A. Chodosh, Room 614 BRB II/III, 421 Curie Boulevard, Philadelphia, Pennsylvania 19104-6160, USA. Email: chodosh@pennmedicine.upenn.edu.
Find articles by Morrissette, J. in: PubMed | Google Scholar
1Secondary Prevention through Surveillance and Intervention (2-PREVENT) Translational Center of Excellence,
2Abramson Family Cancer Research Institute,
3Department of Cancer Biology,
4Department of Pathology and Laboratory Medicine,
5Department of Medicine,
6Department of Radiology, and
7Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Address correspondence to: Lewis A. Chodosh, Room 614 BRB II/III, 421 Curie Boulevard, Philadelphia, Pennsylvania 19104-6160, USA. Email: chodosh@pennmedicine.upenn.edu.
Find articles by Soucier-Ernst, D. in: PubMed | Google Scholar
1Secondary Prevention through Surveillance and Intervention (2-PREVENT) Translational Center of Excellence,
2Abramson Family Cancer Research Institute,
3Department of Cancer Biology,
4Department of Pathology and Laboratory Medicine,
5Department of Medicine,
6Department of Radiology, and
7Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Address correspondence to: Lewis A. Chodosh, Room 614 BRB II/III, 421 Curie Boulevard, Philadelphia, Pennsylvania 19104-6160, USA. Email: chodosh@pennmedicine.upenn.edu.
Find articles by Goodman, N. in: PubMed | Google Scholar
1Secondary Prevention through Surveillance and Intervention (2-PREVENT) Translational Center of Excellence,
2Abramson Family Cancer Research Institute,
3Department of Cancer Biology,
4Department of Pathology and Laboratory Medicine,
5Department of Medicine,
6Department of Radiology, and
7Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Address correspondence to: Lewis A. Chodosh, Room 614 BRB II/III, 421 Curie Boulevard, Philadelphia, Pennsylvania 19104-6160, USA. Email: chodosh@pennmedicine.upenn.edu.
Find articles by Stavropoulos, S. in: PubMed | Google Scholar
1Secondary Prevention through Surveillance and Intervention (2-PREVENT) Translational Center of Excellence,
2Abramson Family Cancer Research Institute,
3Department of Cancer Biology,
4Department of Pathology and Laboratory Medicine,
5Department of Medicine,
6Department of Radiology, and
7Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Address correspondence to: Lewis A. Chodosh, Room 614 BRB II/III, 421 Curie Boulevard, Philadelphia, Pennsylvania 19104-6160, USA. Email: chodosh@pennmedicine.upenn.edu.
Find articles by
Maxwell, K.
in:
PubMed
|
Google Scholar
|

1Secondary Prevention through Surveillance and Intervention (2-PREVENT) Translational Center of Excellence,
2Abramson Family Cancer Research Institute,
3Department of Cancer Biology,
4Department of Pathology and Laboratory Medicine,
5Department of Medicine,
6Department of Radiology, and
7Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Address correspondence to: Lewis A. Chodosh, Room 614 BRB II/III, 421 Curie Boulevard, Philadelphia, Pennsylvania 19104-6160, USA. Email: chodosh@pennmedicine.upenn.edu.
Find articles by Clark, C. in: PubMed | Google Scholar
1Secondary Prevention through Surveillance and Intervention (2-PREVENT) Translational Center of Excellence,
2Abramson Family Cancer Research Institute,
3Department of Cancer Biology,
4Department of Pathology and Laboratory Medicine,
5Department of Medicine,
6Department of Radiology, and
7Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Address correspondence to: Lewis A. Chodosh, Room 614 BRB II/III, 421 Curie Boulevard, Philadelphia, Pennsylvania 19104-6160, USA. Email: chodosh@pennmedicine.upenn.edu.
Find articles by Belka, G. in: PubMed | Google Scholar
1Secondary Prevention through Surveillance and Intervention (2-PREVENT) Translational Center of Excellence,
2Abramson Family Cancer Research Institute,
3Department of Cancer Biology,
4Department of Pathology and Laboratory Medicine,
5Department of Medicine,
6Department of Radiology, and
7Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Address correspondence to: Lewis A. Chodosh, Room 614 BRB II/III, 421 Curie Boulevard, Philadelphia, Pennsylvania 19104-6160, USA. Email: chodosh@pennmedicine.upenn.edu.
Find articles by Feldman, M. in: PubMed | Google Scholar
1Secondary Prevention through Surveillance and Intervention (2-PREVENT) Translational Center of Excellence,
2Abramson Family Cancer Research Institute,
3Department of Cancer Biology,
4Department of Pathology and Laboratory Medicine,
5Department of Medicine,
6Department of Radiology, and
7Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Address correspondence to: Lewis A. Chodosh, Room 614 BRB II/III, 421 Curie Boulevard, Philadelphia, Pennsylvania 19104-6160, USA. Email: chodosh@pennmedicine.upenn.edu.
Find articles by
DeMichele, A.
in:
PubMed
|
Google Scholar
|

1Secondary Prevention through Surveillance and Intervention (2-PREVENT) Translational Center of Excellence,
2Abramson Family Cancer Research Institute,
3Department of Cancer Biology,
4Department of Pathology and Laboratory Medicine,
5Department of Medicine,
6Department of Radiology, and
7Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Address correspondence to: Lewis A. Chodosh, Room 614 BRB II/III, 421 Curie Boulevard, Philadelphia, Pennsylvania 19104-6160, USA. Email: chodosh@pennmedicine.upenn.edu.
Find articles by Chodosh, L. in: PubMed | Google Scholar
Published July 13, 2020 - More info
J Clin Invest. 2020;130(8):4252–4265. https://doi.org/10.1172/JCI129941.
© 2020 American Society for Clinical Investigation
Received: May 8, 2019; Accepted: May 5, 2020
-
Abstract
Nearly all breast cancer deaths result from metastatic disease. Despite this, the genomic events that drive metastatic recurrence are poorly understood. We performed whole-exome and shallow whole-genome sequencing to identify genes and pathways preferentially mutated or copy-number altered in metastases compared with the paired primary tumors from which they arose. Seven genes were preferentially mutated in metastases — MYLK, PEAK1, SLC2A4RG, EVC2, XIRP2, PALB2, and ESR1 — 5 of which are not significantly mutated in any type of human primary cancer. Four regions were preferentially copy-number altered: loss of STK11 and CDKN2A/B, as well as gain of PTK6 and the membrane-bound progesterone receptor, PAQR8. PAQR8 gain was mutually exclusive with mutations in the nuclear estrogen and progesterone receptors, suggesting a role in treatment resistance. Several pathways were preferentially mutated or altered in metastases, including mTOR, CDK/RB, cAMP/PKA, WNT, HKMT, and focal adhesion. Immunohistochemical analyses revealed that metastases preferentially inactivate pRB, upregulate the mTORC1 and WNT signaling pathways, and exhibit nuclear localization of activated PKA. Our findings identify multiple therapeutic targets in metastatic recurrence that are not significantly mutated in primary cancers, implicate membrane progesterone signaling and nuclear PKA in metastatic recurrence, and provide genomic bases for the efficacy of mTORC1, CDK4/6, and PARP inhibitors in metastatic breast cancer.
-
Introduction
Breast cancer is the most prevalent cancer in the United States and the leading cause of cancer mortality worldwide among women (1). Although 5-year survival rates approach 90%, up to 30% of breast cancer patients ultimately die from their disease, typically due to disease recurrence at distant sites following a variable period of clinical remission (2).
Historically, metastatic breast cancer was treated with the tacit assumption that key biological features of primary tumors are preserved during metastatic outgrowth; however, more recent findings challenge this belief. Indeed, the fact that primary breast cancer is frequently curable, whereas metastatic breast cancer is not, suggests that important biological differences exist between these 2 stages of tumor progression.
Because breast cancer mortality is principally due to metastatic disease, improving outcomes for breast cancer patients will be facilitated by a detailed understanding of the biology of metastatic evolution. Unfortunately, while the genomic landscape of primary breast cancer has been extensively analyzed in over 2000 patients (3), analogous data for metastatic breast cancer are relatively sparse.
Studies of recurrent breast cancer in preclinical models and patient cohorts (4–12) have demonstrated that cancers undergo considerable molecular and cellular evolution during the course of tumor recurrence. Consistent with this observation, hormone receptor (HR) and HER2 status are discordant between primary and metastatic tumors in 20% to 25% of patients (13). Further, while most oncogenic driver mutations in primary tumors are retained within metastases — as would be anticipated from their clonal relationship — metastatic tumors harbor additional oncogenic mutations beyond those detected in their primary tumors of origin (8, 10, 12).
Recent studies using targeted sequencing panels in paired primary and metastatic tumors, as well as high-throughput sequencing in unpaired metastases, have identified several genes that appear to be preferentially mutated in breast cancer metastases, including ESR1 (14), ERBB2 (12), JAK2 (10), NF1 (12), PALB2 (6), STAT3 (10), and TSC1/2 (6). However, the majority of metastatic breast cancers do not harbor mutations in these genes, suggesting that others remain to be identified. Moreover, copy number alterations (CNAs) that preferentially occur in metastases, as well as pathways that are preferentially mutated, have yet to be comprehensively characterized. In this study, we sought to identify genes and pathways that are preferentially mutated or copy-number altered within metastases compared with the primary tumors from which they arose in order to elucidate potential drivers of metastasis and therapeutic resistance.
-
Results
Genomic assays and tumor cohort. Whole-exome sequencing (WES) and shallow whole-genome sequencing (sWGS) were performed to detect somatic coding mutations and genome-wide CNAs in paired primary and metastatic tumors from 28 patients and unpaired metastases from 38 additional patients (Supplemental Table 1 and Supplemental Methods; supplemental material available online with this article; https://doi.org/10.1172/JCI129941DS1). Seventy-five percent of recurrent tumors were distant metastases (liver [n = 36], brain [n = 3], lymph node [n = 3], lung [n = 2], soft tissue [n = 2], bone [n = 1], contralateral breast [n = 1], ovary [n = 1], and skin [n = 1]), with the remaining tumors consisting of locoregional recurrences (lymph node [n = 10], chest wall [n = 3], sternum [n = 2], and ipsilateral breast [n = 1]). Primary tumors were predominantly invasive ductal carcinomas (78.8%), with 6.1% exhibiting features of invasive lobular carcinoma and 10.6% exhibiting mixed lobular and ductal features. All genomically assayed primary tumors were treatment naive. Four metastatic tumors (6.1%) were synchronous, defined as having been biopsied within 3 months of primary tumor resection, and arose in treatment-naive patients. The median time between primary tumor resection and asynchronous metastatic tumor biopsy for the remaining samples was 5.7 years (range: 9 months to 27 years). Biopsies of asynchronous metastatic tumors occurred after patients had been exposed to at least 1 form of therapy: chemotherapy (74.2%), radiation therapy (68.2%), antiestrogen therapy (72.7%), and/or anti-HER2 therapy (13.6%).
Metastatic tumors in our cohort arose most commonly from primary tumors that expressed estrogen receptor (ER) and/or progesterone receptor (PR), hereafter referred to as HR+ (75.8%); 12.7% of metastatic tumors arose from HER2-amplified (HER2+) primary tumors, as determined by immunohistochemistry (IHC) and/or fluorescence in situ hybridization (FISH). Considered in combination, metastases in our cohort arose from primary tumors that were HR+HER2– (69.8%), HR–HER2– (triple-negative breast cancer [TNBC], 17.5%), HR+HER2+ (6.3%), or HR–HER2+ (6.3%). Although HR/HER2 status was discordant between primary and metastatic tumors in the same patient in 17.5% of cases, the distribution of HR/HER2 subtypes was similar for primary and metastatic tumors in our cohort (Supplemental Figure 1).
Genomic concordance and germline mutations. Multiregion sequencing (MRS) revealed that metastatic tumors exhibit substantial genomic divergence from the primary tumors from which they arose (Figure 1 and Supplemental Figure 2) and were accompanied by a greater number of coding mutations, CNAs, and large-scale state transitions (LSTs), a “genomic scar” indicative of homologous recombination deficiency (HRD) (15) and BRCA1/2 loss of function (LOF) (16) (see Supplemental Results).
 Figure 1
Figure 1Paired primary-metastatic tumors are genomically distinct. (A) Concordance for coding mutations (Jaccard index) and genome-wide copy number (R2) between primary-metastasis tumor pairs (orange, n = 28), multiple regions assayed from different tissue blocks from the same tumor (purple, n = 14), and sequencing replicates (green, n = 12). Concordance between primary-metastasis tumor pairs is substantially lower than concordance values between multiple regions within the same tumor (1-sided Wilcoxon’s rank-sum test). Dashed vertical lines indicate mean concordance between unrelated tumors. (B) Concordance determined for coding mutations and genome-wide CNs are significantly correlated, R2 = 0.47. Columns represent primary-metastasis tumor pairs. (C) Metastases (M) exhibit increased numbers of somatic coding mutations, CNAs, and LSTs compared with the primary tumors (P) from which they arose (1-sided Wilcoxon’s signed-rank test). Colored lines indicate patients for whom metastatic tumors exhibited an increased (red) or decreased (blue) number of alterations compared with primary tumors from which they arose.
Consistent with prior reports (14), activating mutations in ESR1 (ERα) were detected in metastatic tumors from 7 patients (10.6%), each of whom had been treated with antiestrogen therapy before metastatic tumor biopsy. Mutations in PGR (PR), which are reportedly rare in primary breast cancer and other cancers (17), were identified in 2 metastatic tumors, each of which arose in patients treated with aromatase inhibitors. Four metastases (6.1%) harbored activating mutations in ERBB2 (HER2), each of which was clonal and/or co-occurred with copy number (CN) gain and/or loss of the WT allele. Metastases with ERBB2 mutations arose in both HR–HER2+ and HR+HER2– primary tumors and in anti-HER2– as well as antiestrogen-treated patients.
Ten patients (15%) exhibited germline variants of clinical significance (VCS): BRCA1 (n = 3; E23fs, S1253fs, A1729E), BRCA2 (n = 3; K1036*, W1692fs, K2013*), PALB2 (n = 2; R170fs, Y1108fs), ATM1 (n = 1; V1602fs), and CHEK2 (n = 1; T410fs). All identified BRCA1, BRCA2, and PALB2 germline variants were confirmed by targeted panel sequencing in a CLIA-certified laboratory. The frequency of HRD-associated LSTs was increased in metastases that harbored at least 1 germline VCS in BRCA1, BRCA2, or PALB2 (P = 0.024), which is consistent with their roles as inherited breast cancer susceptibility genes that promote homologous recombination repair (18).
All but 3 germline variants (BRCA2 K2013*, PALB2 R170fs, and PALB2 T1108fs) co-occurred with loss of heterozygosity (LOH) in their respective tumors. Both patients with germline PALB2 mutations developed tumors harboring additional clonal somatic PALB2 mutations, likely leading to loss of the WT allele and complete LOF. Furthermore, 3 metastases in patients carrying WT germline alleles for BRCA1 and BRCA2 exhibited clonal somatic mutations in BRCA1 or BRCA2 that co-occurred with LOH or mutation of the remaining allele. In total, 15% (n = 10) of metastases were predicted to have complete LOF of BRCA1, BRCA2, or PALB2.
Significantly mutated genes in metastases. Fifteen significantly mutated genes (SMGs) were identified in our metastatic tumor cohort using MutSigCV2 (19). Seven SMGs are not significantly mutated in primary breast cancers in The Cancer Genome Atlas (TCGA-BRCA), either within or across subtypes: ESR1, XIRP2, PEAK1, PALB2, SLC2A4RG, MYLK, and EVC2. Five of these have not been reported as SMGs by TCGA in any type of primary cancer: ESR1, MYLK, PEAK1, SLC2A4RG, and EVC2. An additional 8 SMGs have previously been reported in TCGA-BRCA: PIK3CA, TP53, KMT2C/MLL3, GATA3, RUNX1, CHD1, MAP2K4, and AKT1.
To determine whether the above 15 SMGs were preferentially mutated in treatment-refractory metastases, we compared their mutational frequencies to those in primary breast cancers from TCGA-BRCA (n = 1043). Mutations in TCGA-BRCA were first recalled using the same variant-calling pipeline and stringency threshold criteria used in our study (see Methods). Each of the 7 SMGs identified in our cohort that are not SMGs in primary breast cancer, as well as RUNX1, exhibited significantly higher mutation frequencies in metastases compared with TCGA-BRCA primary tumors using both the high-confidence thresholds employed in this study and lower-confidence thresholds employed in the TCGA-BRCA study (FDR ≤ 0.10; Figure 2A, Supplemental Table 2, and ref. 3). These differences in mutation frequencies persisted when restricting the analysis to TCGA-BRCA samples with the highest gene coverage for each SMG (Supplemental Figure 3) and were not explained by differences in receptor subtype distributions between the 2 data sets (Supplemental Figure 1). Increased mutational frequencies were also observed for TP53, KMT2C, and AKT1; however, these differences were not significant after accounting for differences in coverage.
 Figure 2
Figure 2Genes preferentially mutated in metastases. (A) Mutation frequencies for SMGs identified by MutSigCV2 within metastatic tumors in our cohort (red, n = 66) and primary tumors in TCGA-BRCA (blue, n = 1044). Seven SMGs, indicated in bold, have not been reported in TCGA-BRCA primary tumors across nor within subtypes. Eleven SMGs exhibited significantly higher mutation frequencies in metastases within our cohort compared with TCGA-BRCA primary tumors (2-sided Fisher’s exact test; *FDR ≤ 0.10; **FDR < 0.001). Red and orange asterisks respectively denote 3 SMGs that either lose or gain significance when less stringent filtering criteria employed in Ciriello et al. (3) are used. (B) Co-occurrence of MYLK and PEAK1 mutations with ERBB2 mutations and HER2+ status in metastases (2-sided Fisher’s exact test, *P < 0.05; **P < 0.01; FDRs = 0.07–0.36). Each column represents a metastatic tumor. (C) Kaplan-Meier survival analysis showing that TCGA-BRCA patients whose primary tumor had a mutation in MYLK exhibited shorter RFS.
Interestingly, 3 SMGs involve the regulation of actin polymerization downstream of FAK signaling: the myosin light-chain kinase MYLK (or MLCK) (20), the actin-binding protein XIRP2 (21), and the nonreceptor tyrosine kinase PEAK1 (22). Mutations in PEAK1 were either nonsense (n = 2) or co-occurred with CN loss (n = 3), and recurrences generally exhibited lower PEAK1 copy number (P = 0.035), suggesting that PEAK1 mutations in metastases are inactivating. SLC2A4RG is a transcriptional activator of the glucose transporter SLC2A4 (GLUT4) (23). Mutations in SLC2A4RG in metastases co-occurred with CN gain, suggesting that they may be activating. However, co-occurrence with CN gain may be incidental due to the proximity (~200 kbp) of SLC2A4RG to the putative CN driver, PTK6 (see below). Germline LOF mutations in PALB2 result in inherited breast cancer susceptibility; however, PALB2 is not an SMG in primary breast cancer. EVC2 encodes a transmembrane protein reported to be hypermethylated in neoadjuvant treatment–resistant TNBC (24). LOF mutations in this gene result in the inherited skeletal dwarfism disorder Ellis–van Creveld (EvC) syndrome and have been implicated in defective hedgehog signaling (25) as well as elevated fibroblast growth factor signaling (26).
Mutations in MYLK and PEAK1 preferentially occurred in HER2+ metastases (P = 0.0017, FDR = 0.04 and P = 0.0078, FDR = 0.13, respectively; Figure 2B) and in ERBB2-mutant metastases (P = 0.016, FDR = 0.23 and P = 0.039, FDR = 0.36, respectively), with 1 HER2+ metastatic tumor exhibiting mutations in ERBB2, MYLK, and PEAK1. Mutations in these genes are not correlated with HER2 status in primary breast cancers in TCGA-BRCA (P > 0.40), suggesting that these associations may be specific to the metastatic, treatment-refractory setting. Consistent with this, relapse-free survival (RFS) was reduced in TCGA-BRCA patients whose primary tumors harbored mutations in MYLK (HR+HER2– tumors, P = 3.3 × 10–4, FDR < 0.01; all tumors, P = 0.0038, FDR = 0.05; Figure 2C). No significant associations with RFS were found for other metastasis-enriched SMGs, although statistical power was limited by the rarity of these mutations in primary tumors.
Preferential focal CNAs in metastases. GISTIC2 (27) was used to identify focal genomic regions with significantly increased frequencies of CNAs in either primary or metastatic tumors (FDR ≤ 0.10; Supplemental Table 3). Whereas all significantly altered regions (SARs) recurrently deleted (n = 20) in metastases were reported by TCGA-BRCA within at least 1 breast cancer subtype, the majority of recurrently amplified SARs (11 of 17) were unique to this metastatic tumor cohort, including 1q32.1-q32.2, 3q29, 6p12.2, 6q25.1-q25.2, 7p22.1-21.3, 10q21.2, 14p13-q11.1, 15p11.1-q11, 17q21.1, 17q21.31, and 20p11.1-q11.21.
Two SARs were preferentially lost and/or exhibited decreased CN in metastases compared with their primary tumor of origin: 19p13.3 (FDR = 0.03) and 9p21 (FDR = 0.13; Figure 3, A and B, and Supplemental Table 3). Although 44 genes reside in 19p13.3, STK11/LKB1 is located at its center and has previously been implicated as an oncogenic driver for loss of this region (28). STK11 loss is predicted to downregulate AMPK and upregulate mTOR signaling (29), is associated with poor prognosis in breast cancer, and has been implicated in metastasis in a variety of cancers (30). Recurrent deletion of 9p21 occurs in a broad spectrum of human cancers, including primary breast cancer (31), and has been ascribed to loss of CDKN2A (encoding p14ARF and p16INK4A) and CDKN2B (encoding p15INK4B).
 Figure 3
Figure 3Copy number alterations enriched in metastases. (A) Frequencies of CN gain (red) and CN loss (blue) within primary (P, n = 28) and metastatic (M, n = 66) tumors for 4 SARs preferentially altered in metastases. Low-level CNAs (gain and loss) and high-level CNAs (amplification and deletion) are shown as lighter- and darker-colored bars, respectively. χ2 P values indicate the degree to which the distributions of low-level CNA frequencies are different between primary and metastatic tumors. Putative driver genes for each SAR are indicated. (B) Normalized CN values for paired primary and metastatic tumors (2-sided Wilcoxon’s signed-rank test). (C) Kaplan-Meier survival analysis within the METABRIC data set (n = 1483) showing association of 3 of 4 identified metastasis-enriched SARs with shorter DSS. (D) Gain of PAQR8 in metastases is mutually exclusive with mutations in ESR1 or PGR in patients treated with antiestrogen therapy (2-sided Fisher’s exact test). Each column represents a metastatic tumor.
Two SARs were preferentially gained and/or exhibited increased CN in metastases compared with their primary tumor of origin: 6p12.2 (FDR = 0.09) and 20p11.1-q11.21 (FDR = 0.07). The 78-kbp SAR within 6p12.2 contains only a single gene: the membrane-bound PR, PAQR8 (mPRβ). mPRs have been reported to mediate rapid, nongenomic effects of progesterone and to regulate cyclic adenosine monophosphate (cAMP) synthesis (32) and apoptosis (33). Strikingly, in metastases arising in patients treated with antiestrogen therapy, PAQR8 gain was mutually exclusive with mutations in either ESR1 or PGR (66.7% of ESR1/PGR-WT metastases exhibited PAQR8 CN gain compared with 11.1% of ESR1/PGR mutant metastases, P = 0.0062; ESR1 alone, P = 0.032; Figure 3D). Nevertheless, frequencies of PAQR8 gain in metastases were similar in patients who had, or had not, been treated with antiestrogen therapies (56% in each). These findings suggest that PAQR8 gain may contribute to resistance to endocrine therapies as well as chemotherapy.
The preferential gain of 20p11.1-q11.21 observed in metastases was notable because chr20q was amplified in its entirety in the majority of metastases in our cohort (57.6%, GISTIC2 FDR < 0.01). Because 20p11.1-q11.21 closely corresponds to the gene-poor pericentromeric region of this chromosome, we considered that this SAR may simply represent a marker for increased chr20q CN in metastases compared with primary tumors (arm-level CN difference P = 0.0065, FDR = 0.09). Within this chromosome arm, 20q13.33 exhibited the largest CN difference between paired primary and metastatic tumors (P = 9.1 × 10–4, FDR = 0.03 when included as a GISTIC2-identified SAR; Supplemental Figure 4). 20q13.33 contains the nonreceptor tyrosine kinase PTK6 (BRK, breast tumor kinase), which is amplified and overexpressed in a variety of human cancers (34). PTK6 has been reported to promote proliferation, survival, and metastasis (35), as well as resistance to targeted therapies (36), in part through its interaction with receptor tyrosine kinases (RTKs) such as EGFR and ERBB2 (37).
Within the METABRIC data set (38), disease-specific survival (DSS) was significantly shorter in patients whose primary tumors harbored loss of CDKN2A, loss of STK11, or gain of PTK6 (P = 2.5 × 10–11, 0.0011, 0.0021, respectively; Figure 3C). This relationship also held for ER+ tumors alone (P = 4.0 × 10–10, 6.9 × 10–4, 0.0063, respectively). PAQR8 gain was also associated with decreased DSS, but only within the METABRIC CN cluster subtype, IntClust7 (P = 0.0020), which is a subgroup of luminal A tumors. Because these associations cannot be attributed to increased CNA burden in metastases (see Supplemental Results), these observations provide evidence for a role for CDKN2A loss, STK11 loss, PTK6 gain, and PAQR8 gain in promoting metastasis and/or treatment resistance.
Pathways preferentially mutated in metastases. Patient-oriented gene set enrichment analysis (39) identified 7 significantly mutated KEGG-defined pathways (SMPs) in primary tumors, 43 SMPs in paired metastatic tumors, and 58 SMPs across all metastatic tumors (FDR ≤ 0.10). For 30 SMPs, significantly more patients exhibited private mutations (i.e., mutations not shared between paired tumors, see Methods) exclusively in the metastatic tumor and few, if any, patients had private mutations solely in the primary tumor (FDR ≤ 0.10, Pperm ≤ 0.10; Supplemental Table 4). Twelve of these SMPs (mTOR Signaling, PI3K-AKT Signaling, Focal Adhesion, Progesterone-mediated Oocyte Maturation, cAMP Signaling, Lysine Degradation, Regulation of Lipolysis in Adipocytes, Longevity Regulating, VEGF Signaling, Prolactin Signaling, HIF-1 Signaling, and Carbohydrate Digestion and Absorption) were also preferentially mutated in 211 metastatic tumors from Lefebvre et al. (6) compared with 1,044 TCGA-BRCA primary tumors (Figure 4A, Supplemental Table 5, and see Methods), which constitutes a significant overlap (P = 2.0 × 10–6). Recurrently mutated genes and modules within these 12 metastasis-enriched SMPs are reported in the Supplemental Results, though not all of these mutations necessarily modulate pathway activity.
 Figure 4
Figure 4Pathways preferentially mutated in metastases. Thirty pathways that were preferentially mutated in metastases compared with paired primary tumors. (A) SMPs identified in the current study as being enriched in metastases from this cohort as well as in metastases from Lefebvre et al. (6) compared with TCGA-BRCA primary tumors. (B) SMPs enriched in metastases in this cohort, but not enriched in Lefebvre et al. compared with TCGA-BRCA. Significance in the local cohort was determined by comparing the fraction of primary-metastasis tumor pairs in which only the primary tumor has a private mutation (blue), only the metastatic tumor has a private mutation (red), or both primary and metastatic tumors have at least 1 private mutation (purple) (McNemar’s test; FDR < 0.10). Permutation analysis was used to control for the global increase in coding mutations within metastases (*Pperm < 0.10, **Pperm < 0.05, ***Pperm < 0.01).
Eighteen metastasis-enriched SMPs were identified in our primary/metastatic tumor data sets, but not in the Lefebvre/TCGA-BRCA data sets (Figure 4B): Estrogen Signaling, Wnt Signaling, Phospholipase D Signaling, Inflammatory Mediator Regulation of TRP Channels, Toll-like Receptor Signaling, Platelet Activation, Long-term Depression, Protein Digestion and Absorption, Axon Guidance, Rap1 Signaling, Relaxin Signaling, Fluid Shear Stress and Atherosclerosis, Fc Epsilon RI Signaling, Leukocyte Transendothelial Migration, Neurotrophin Signaling, Sphingolipid Signaling, Autophagy-Animal, and TNF signaling.
mTOR is hyperactivated in metastases. Metastases preferentially harbored mutations in the mTOR signaling pathway both within and beyond core RTK/PI3K/AKT pathway components (Supplemental Figures 5 and 6). Of particular interest were mutations in the amino acid–sensing subpathway (e.g., vATPase and GATOR1/2 complexes), WNT receptors and ligands (in which mutations were only present in metastases), the MAPK pathway (in which all mutations were predicted to be activating), TSC1/2 core regulators, and subunits of mTORC1/2. Though not previously reported in cancer, a mutation in the mTORC2 subunit RICTOR, K1125E, is structurally similar to an acetylation mimic, K1125Q, that constitutively activates mTORC2 independently of glucose levels (40). Overall, mutations occurring in the mTOR pathway in metastases were consistent with mTOR activation in 26% of cases, compared with only 9% of cases in which mutations suggested mTOR inactivation.
Beyond these mutations, the frequent and preferential loss of STK11 and gain of PTK6 observed in metastases would also be predicted to activate mTOR by inhibiting AMPK (29) or by coactivating RTKs (37), respectively. Strikingly, STK11 loss was present in every metastasis that arose from a primary tumor bearing a non-PIK3CA mutation in the mTOR Signaling gene set (P = 0.0047, FDR = 0.12; Supplemental Figure 5). Furthermore, STK11 loss co-occurred with PTK6 gain in metastases (P = 0.038, FDR = 0.20). This suggests that multiple alterations predicted to activate mTOR may be coselected within treatment-refractory metastases.
IHC was performed to evaluate levels of phosphorylated ribosomal protein S6 (p-S6), a functional read-out of mTOR activity (41), in 2 cohorts: 15 paired primary and metastatic tumors that arose within the same patient, and 16 matched sets of primary and metastatic tumors that arose in different patients. In the matched cohort, primary and metastatic tumors were matched for tumor block age and preservation type, as well as the receptor subtype of the primary tumor from which the metastasis arose (Figure 5).
 Figure 5
Figure 5mTOR is preferentially activated in metastases. (A) Representative p-S6 IHC images for primary-metastasis tumor pairs from the same patient (n = 15), and primary and metastatic tumors matched for tumor block age, preservation type, and receptor subtype of the primary tumor from which the metastasis arose (n = 16). p-S6 IHC intensity scoring scale (no p-S6 signal = 0, saturated signal = 3), with isotype control. (B) Mean p-S6 intensity and (C) proportion of cells that are p-S6+ in paired and matched primary (P) and metastatic (M) tumors (1-sided Wilcoxon’s signed-rank test). (D) Mean p-S6 intensity and (E) proportion of cells that were p-S6+ (score = 3) in metastases WT or mutant for genomic features that either exhibited a significant (1-sided Wilcoxon’s signed-ranked test, P < 0.05, white) or nonsignificant (gray) association with p-S6 intensity. Gene names refer to genes that are mutated, unless in reference to low-level CN gain or loss. “mTOR (≥x)” indicates the presence of at least x number of non-PIK3CA mutations in the mTOR Signaling pathway.
Both mean p-S6 staining intensity (Figure 5B) and the proportion of tumor cells that were p-S6+ (Figure 5C) were significantly higher in metastatic tumors than in their corresponding primary tumors (median increase in intensity = 2.0-fold [IQR = 1.3–7.9], P = 9.8 × 10–5; proportion of cells with p-S6 score = 3, P = 2.9 × 10–6; with score ≥ 2, P = 1.8 × 10–4; with score ≥ 1, P = 0.0039). Differences in mTOR activity remained significant when paired and matched tumor cohorts were analyzed separately (Supplemental Figure 7, A and B). Importantly, p-S6 staining within the tumor stroma, nontumor stroma, and nontumor epithelium were not significantly different between primary tumors and metastases, suggesting that observed differences in p-S6 staining in tumor cells were not attributable to systematic technical differences in processing of primary tumors and metastatic biopsies, such as time from excision to fixation (Supplemental Figure 7C).
Compared with their corresponding primary tumors, mTOR activity was elevated in both liver and nonliver metastases (P = 0.013 and 0.0012, respectively; Supplemental Figure 7D), and irrespective of whether patients were treated with antiestrogen therapy or not (P = 0.001 and 0.020, respectively; Supplemental Figure 7E). Together, these genomic and biochemical findings demonstrate that mTOR is hyperactivated in metastases compared with primary tumors.
Genomic alterations associated with mTOR activity. Genomic alterations in the mTOR pathway were evaluated for their association with p-S6 staining intensity in metastases (Supplemental Figure 8). Surprisingly, neither PIK3CA mutation status nor PTEN loss was associated with p-S6 intensity in metastases or primary tumors (Supplemental Figure 9, A and B). In contrast, mean p-S6 intensity was significantly higher in metastases that harbored 2 or more mutations in the mTOR Signaling pathway (P = 0.019), irrespective of PIK3CA mutation status (Supplemental Figure 9C). Three genomic alterations known to activate mTOR signaling were significantly associated with increased p-S6 staining (Figure 5, D and E): ERBB2 mutation (P = 0.0070), HER2+ status (P = 0.038), and loss of STK11 (P = 0.040). CNAs in 3 additional genes not known to affect mTOR pathway activity were also associated with increased p-S6 staining: gain of PAQR8 (P = 0.018), gain of CCNE1 (P = 0.014), and loss of TP53 (P = 0.011). For each of these alterations, p-S6 was elevated in both mutant and WT metastases compared with genomically assayed paired primary tumors (Supplemental Table 6). Notably, the 4 metastases with weakest p-S6 intensity all lacked the presence of any genomic alteration associated with mTOR activity. Intriguingly, 3 of these metastases harbored mutations in PIK3CA (n = 2) and/or loss of PTEN (n = 2). For 2 of these metastases whose paired primary tumor was assayed by p-S6 staining, p-S6 intensity did not differ between the primary and metastasis.
pRB is preferentially inactivated in metastases. In light of the frequent and preferential loss of CDKN2A in metastases, we estimated the extent to which pathways impinging on pRB were dysregulated (Figure 6A). Three metastases exhibited clonal mutations in CDKN2A (R80*, P81A, and D84N) within a mutational hotspot known to abrogate the ability of p16INK4A to bind and inhibit CDK4 and CDK6 (42). Further, the R80* mutation was estimated to be the only CDKN2A allele present, suggesting that p16INK4A was entirely inactivated in this metastasis. Three additional metastases harbored mutations in RB1 (n = 2) or RBL2 (RB2/p130), each of which was a frameshift mutation that was clonal and co-occurred with CN deletion. These mutational data suggest that pRB may be dysregulated in metastases.
 Figure 6
Figure 6pRB is preferentially inactivated in metastases. (A) Pathway schematic of genes downstream of p16INK4A/p14ARF and/or upstream of pRB. Solid and dashed lines indicate direct and indirect relationships between gene products. The frequencies of high-level CNAs (upper) and mutations (lower, where provided) in each gene are shown for primary tumors (left) and metastases (right) beneath each gene. (B) Representative phospho-RB IHC images for paired (n = 8) and matched primary and metastatic tumors (n = 15). (C) Mean phospho-RB staining intensity and (D) proportion of cells that are phospho-RB+ in paired and matched primary and metastatic tumors (1-sided Wilcoxon’s signed-rank test).
Consistent with the possibility of pRB inactivation in metastases, nearly twice the number of high-level CNAs predicted to inactivate pRB were identified in metastatic tumors compared with primary tumors (mean, primary = 1.1 ± 1.0, metastasis = 2.1 ± 1.7; P = 0.005), including gains of CCNB1, CCND1, CCNE1, CCNH, CDK1, CDK2, CDK4, CDK6, CDK7, and MDM2, as well as losses of CDKN2A, TP53, and RB1 (Supplemental Figure 10). This imbalance is unlikely to result from global increases in CNA frequencies in metastases, since the overall number of pRB-activating CNAs observed in primary and metastatic tumors were similar (mean, primary = 0.8 ± 1.6, metastasis = 0.6 ± 1.2; P = 0.55).
To determine whether preferential inactivation of the CDK/RB pathway in metastases occurred at the biochemical level, IHC was performed for phosphorylated pRB (phospho-RB), a functional read-out of pRB inhibition, in a subset of tumors from the same cohort assayed for p-S6 activity (Figure 6B). Compared with their corresponding primary tumors, metastases exhibited significantly higher mean phospho-RB intensities within the tumor epithelium (median of 4.2-fold increase [IQR = 1.8–7.7], P = 5.1 × 10–4), as well as higher proportions of cells that were phospho-RB+ (Figure 6, C and D). phospho-RB mean intensities were significantly higher in metastases than primary tumors in both paired (n = 8) and matched (n = 15) tumor cohorts, liver and non-liver metastases, and patients treated — or not treated — with antiestrogen therapy (Supplemental Figure 11). Together, genomic and biochemical analyses indicate that pRB is preferentially inactivated in metastatic tumors.
Higher percentages of cells with saturated phospho-RB signal were observed in metastases harboring a mutation in ERBB2 or loss of TP53 (score = 3; P = 0.032 and 0.067, respectively). In addition, percentage positivity (score ≥ 1) and mean phospho-RB intensities trended lower in metastases harboring fewer copies of RB1 (P = 0.088 and 0.13, respectively). Indeed, the metastasis with the lowest mean phospho-RB intensity was the only IHC-assayed metastasis that harbored both a frameshift mutation in RB1 and RB1 deletion, underscoring the inability of phospho-RB IHC to detect all forms of RB inactivation. Nevertheless, even after normalizing for RB1 CN, no significant associations were found between genomic alterations and levels of phospho-RB.
Concomitant alterations in mTOR and CDK/RB pathways. Mutations and CNAs in the mTOR and CDK/RB pathways significantly co-occurred in metastases (Supplemental Figure 12). CN values for CDKN2A in primary tumors were significantly associated with CN values for STK11 in the metastases to which they gave rise (P = 1.9 × 10–4, FDR = 0.02). The reverse was true as well, with CN values for STK11 in primary tumors being significantly associated with CN values for CDKN2A in the metastases to which they gave rise (P = 0.0013, FDR = 0.07). In an analogous manner, patients whose primary tumors harbored a non-PIK3CA mutation in the mTOR pathway gave rise to metastatic tumors with fewer copies of CDKN2A (P = 0.0066, FDR = 0.15). More broadly, high-level CNAs predicted to activate mTOR significantly co-occurred with those predicted to inactivate pRB (P = 2.7 × 10–4, Supplemental Figure 12D), and mean p-S6 and phospho-RB staining intensities within metastases showed some correlation (n = 23, P = 0.13). In aggregate, integrative genomic analysis indicates that mTOR pathway activation and RB pathway inactivation co-occur in metastases, suggesting that alterations in these pathways may cooperate during metastatic recurrence following therapy.
WNT is preferentially activated in metastases. Metastases preferentially harbored mutations in the WNT Signaling gene set, primarily within the canonical WNT signaling pathway (Supplemental Figure 13). IHC was performed to evaluate expression of β-catenin, a main effector of canonical WNT signaling, in primary and metastatic tumors (n = 15, 7 paired and 8 matched sets, Figure 7, A–C).
 Figure 7
Figure 7Preferential WNT activation and nuclear localization of activated PKA in metastases. IHC analyses of β-catenin (A–C) and p-PKA (D–F) in primary (P) and metastatic (M) tumors (n = 15). (A) Representative β-catenin IHC images for primary and metastatic tumors. (B) Mean membranous β-catenin staining intensity and (C) proportion of nuclei that are β-catenin–positive in paired and matched primary and metastatic tumors (1-sided Wilcoxon’s signed-rank test). (D) Representative p-PKA IHC images for primary and metastatic tumors. (E) Mean cytoplasmic p-PKA staining intensity and (F) proportion of nuclei that are p-PKA–positive in paired and matched primary and metastatic tumors.
Activation of canonical WNT signaling may be accompanied by increased β-catenin in the nucleus and decreased β-catenin at the cell membrane, where β-catenin is normally sequestered by E-cadherin. Whereas most primary tumors (n = 13, 86%) exhibited some membranous β-catenin staining, relatively few (n = 4, 27%) exhibited nuclear staining. Metastases exhibited increased staining for both membranous (P = 0.0013, primary vs. metastasis median intensity = 1.70 vs. 2.70) and nuclear β-catenin (P = 0.038, primary vs. metastasis median intensity = 0.00 [max = 0.03] vs. 0.01 [max = 0.23]), with a larger proportion of metastases exhibiting some nuclear β-catenin expression (P = 0.033, n = 10 [66.7%]). Further, nuclear β-catenin expression was observed in metastases in each organ site tested: lymph node (n = 2, max = 0.12), liver (n = 11, max = 0.15), and brain (n = 2, max = 0.23). Overall, β-catenin levels in the membrane and nuclear compartments of metastases (but not primary tumors) were negatively correlated (P = 0.046, R2 = 0.27). No significant associations were found between β-catenin expression and specific genomic alterations. Together, these data suggest that canonical WNT signaling may be preferentially activated in metastases.
Preferential nuclear localization of p-PKA in metastases. Five metastasis-enriched pathways, cAMP Signaling, Progesterone-mediated Oocyte Formation, Longevity Regulating, Carbohydrate Digestion and Absorption, and Regulation of Lipolysis in Adipocytes share core PI3K/AKT/mTOR and cell cycle components with other metastasis-enriched SMPs. Specific to these pathways, however, are genes that regulate the synthesis and action of cAMP (Supplemental Figure 14). cAMP serves as a second messenger in a broad array of cellular processes, including apoptosis, metabolism, differentiation, and proliferation, many of which are mediated through activation of the cAMP-dependent protein kinase, PKA (43).
Mutations in the cAMP Signaling pathway were significantly enriched in metastases and occurred more frequently in metastases from patients who had been treated with either chemotherapy or radiation therapy (P = 2.5 × 10–4). cAMP/PKA is the core effector of the metastasis-enriched Progesterone-mediated Oocyte Formation pathway, which is thought to be driven by nongenomic G protein–coupled mPRs, of which PAQR8 is a prominent member that undergoes preferential CN gain in metastases (see above).
Phosphorylation of threonine 197 within the activation loop of the catalytic subunit of PKA converts this enzyme to an active state. IHC was performed to evaluate expression of this phosphorylated, activated form of PKA (p-PKA T197) within primary and metastatic tumors (n = 15; 7 paired and 8 matched sets; Figure 7, D–F). Cytoplasmic p-PKA was present in all tumors assayed and did not differ between primary and metastatic tumors (median intensity, primary = 1.85 [IQR = 1.32–1.99], metastasis = 1.85 [IQR = 1.05–1.96]) (Figure 7E). In contrast, the proportion of nuclei staining positive for p-PKA was substantially and significantly higher in metastases compared with primary tumors (P = 8.1 × 10–4; median proportion, primary = 1% [IQR = 0%–2%], metastasis = 11% [IQR = 6%–35%]) (Figure 7F). Nuclear p-PKA was observed in more than 1% of tumor cells in only 4 primary tumors (27%), but was detected in more than 1% of tumor cells in all metastases analyzed (n = 15, P = 2.5 × 10–5), and across all organ sites tested: liver (n = 11), brain (n = 2), and lymph node (n = 2). No genomic alterations were identified that were significantly associated with the presence of nuclear p-PKA. The preferential nuclear localization of p-PKA (T197) in metastases suggests that cAMP/PKA signaling is preferentially altered in metastases and may contribute to metastasis and/or the acquisition of treatment resistance.
Preferential mutation of the focal adhesion pathway. Frequent mutations were identified in the Focal Adhesion pathway (Supplemental Figure 15), the majority of which occurred in collagens (26% of metastases harbored mutations in 14 members), integrins (15% of metastases harbored mutations in 9 members), and laminins (11% of metastases harbored mutations in 7 members). Mutations were also identified in a variety of downstream genes regulating the actin cytoskeleton, including the metastasis-enriched SMGs, MYLK, XIRP2, and PEAK1. Non-PIK3CA mutations in the Focal Adhesion pathway were common and occurred at similar frequencies in patients who had, or had not, been treated with antiestrogen therapy (75% and 78%, respectively).
FAK activation inhibits pRB via cyclin D1 (44) and activates mTORC1 (45) as a consequence of shared signaling modules between the Focal Adhesion pathway and upstream activators of mTORC1. Interestingly, 38% of metastases harbored unique mutations in both the mTOR and Focal Adhesion pathways and Focal Adhesion–mutant primary tumors were more likely to give rise to metastatic tumors with STK11 loss (P = 0.0013, FDR = 0.07) (Supplemental Figure 15, B and C). These observations suggest that concomitant dysregulation of the Focal Adhesion and mTOR pathways may be selected for during metastatic outgrowth and/or contribute to treatment resistance.
Preferential mutation of histone lysine methyltransferases. The majority of mutations within the metastasis-enriched SMP, Lysine Degradation, occurred in SET domain–containing histone lysine methyltransferases (HKMTs). Mutations occurred most frequently within the KMT2 gene family, which methylates H3K4, a marker of transcriptionally active chromatin. Metastases exhibited a significantly higher frequency of mutations in KMT2C compared with TCGA-BRCA when confidence thresholds employed in the TCGA-BRCA study were used (Figure 2A). Mutations were also frequently identified in the KMT1, KMT3, KMT5, and KMT8 gene families. Of note, mutations in HKMTs were enriched in metastases harboring somatic mutations in BRCA1, BRCA2, or PALB2 somatic mutations (P = 0.0049), but not in metastases harboring germline VCS in BRCA1, BRCA2, or PALB2 (P = 0.25). The preferential mutation of HKMTs in metastases suggests the potential involvement of epigenetic dysregulation in metastatic outgrowth and/or treatment resistance.
Associations between metastatic sites and genomic alterations. Metastasis-enriched SMGs, SARs, and SMPs were evaluated to determine if their presence was more likely to occur in metastases at specific sites. Compared with metastases at other sites, liver metastases were enriched for HR+ tumors (P = 0.0035), exhibited lower STK11 CN (P = 0.022), and were less likely to harbor PTK6 gain (P = 0.041). Moreover, compared with metastases at other sites, brain metastases exhibited higher PAQR8 CN (P = 0.016) and bone metastases were less likely to exhibit mutations in the metastasis-enriched Focal Adhesion (P = 0.012), cAMP Signaling (P = 0.029), and mTOR Signaling (P = 0.044) SMPs. However, these associations did not retain significance following multiple-test correction.
-
Discussion
Breast cancer mortality is principally due to metastatic recurrence; however, little is known about the biological differences between primary and metastatic tumors in patients. In this study, WES and sWGS were performed on paired primary and metastatic tumors from 28 patients and 38 additional metastases to elucidate genomic events that occur during tumor progression. A principal conclusion of this study is that metastatic tumors are biologically distinct from the primary tumors from which they arise. Specifically, we found that metastases exhibit markedly divergent mutational landscapes, frequently manifest a different ER/PR/HER2 status, acquire additional oncogenic mutations, and exhibit increased genomic instability (Supplemental Figure 16). Most notably, nearly half of the SMGs identified in this study were more frequently mutated in metastases than in primary tumors and have not been reported as SMGs in primary breast cancers: MYLK, PEAK1, XIRP2, EVC2, SLC2A4RG, PALB2, and ESR1. Indeed, 5 of these SMGs (MYLK, PEAK1, EVC2, SLC2A4RG, and ESR1) are not SMGs in any type of primary human cancer in TCGA. In an analogous manner, of the 4 SARs identified as preferentially altered in metastases (loss of STK11 and CDKN2A, gain of PAQR8 and PTK6), 2 (containing PAQR8 and PTK6) have not been reported in primary breast cancers in TCGA. Together, our findings suggest that these 7 SMGs and 2 SARs, along with preferential alterations in the mTOR, CDK/RB, cAMP/PKA, WNT, focal adhesion, and HKMT pathways, contribute to aspects of metastasis that are not rate limiting for primary tumorigenesis and/or represent pathways of therapeutic escape.
Integrated genomic analysis coupled with biochemical validation demonstrated robust hyperactivation of mTOR signaling in metastases compared with primary tumors — a finding bolstered by the observation that STK11 is preferentially lost, and PTK6 preferentially gained, in metastases. mTOR pathway activation has been implicated as a mechanism of escape from therapies that target ER and HER2 in preclinical models (46) and in breast cancer patients (47). Furthermore, treatment with the mTORC1 inhibitor everolimus improved progression-free survival in patients with HR+HER2– metastatic breast cancer when administered along with an aromatase inhibitor (48). Consistent with our findings, a reverse-phase protein microarray analysis identified AKT/mTOR activation in liver metastases compared with unmatched primary breast cancers (49). Unlike these prior findings, however, we observed increased mTOR activity in both liver and nonliver metastases. Similarly, whereas mTOR has been reported to be preferentially activated in breast cancer metastases in patients who received adjuvant endocrine therapy (47), we also observed increased mTOR activity in metastases from patients who had not been exposed to endocrine therapy. In aggregate, our data provide a genomic basis for the efficacy of mTOR inhibition in patients with advanced breast cancer and further suggest that mTOR hyperactivation in metastases occurs in contexts beyond those associated with antiestrogen therapy or metastatic spread to the liver.
Mutational activation of PIK3CA is one of the most common oncogenic drivers of HR+ primary breast cancer (50) and is generally presumed to activate mTORC1 via AKT-mediated inactivation of TSC1/2. It is therefore notable that the increased frequency of mutations that we observed in the mTOR pathway typically involved mTOR pathway components other than PI3K and AKT, including those within the WNT, MAPK, and amino acid–sensing pathways. This suggests that mutations within these pathways are nonredundant with PIK3CA and that activating mutations in PIK3CA may not maximally activate mTORC1. Consistent with this, p-S6 staining was not associated with PIK3CA mutation status, nor PTEN loss, but was instead associated with ERBB2 mutations, HER2+ status, loss of STK11, and the number of non-PIK3CA mutations in the mTOR pathway. Together, our results suggest that PIK3CA mutations alone do not result in sufficient activation of mTORC1 to enable metastasis and/or confer therapeutic resistance in breast cancer, and that multiple genomic alterations in this pathway may be required.
In this study, preferential inactivation of pRB in breast cancer metastases was first suggested by integrative genomic analyses and then confirmed by IHC. Preferential inactivation of pRB was observed irrespective of metastatic site (liver or non-liver), or exposure to endocrine therapy. In light of evidence that increased CDK4/6 activity in ER+ tumors is a demonstrated mechanism of resistance to endocrine therapies (51), and that dual inhibition of CDK4/6 and ER confers improved outcomes in patients with advanced breast cancer (52), our findings provide a genomic basis for understanding the efficacy of CDK4/6 inhibitors in endocrine treatment–refractory metastases.
Of note, the increased frequencies of CCNE1, CDK1, and CDK2 amplification observed in metastases suggest that pRB inactivation in metastases may occur via activation of CDK1/2 as well as CDK4/6. If true, this would imply that CDK4/6 inhibition alone may not achieve durable responses in patients. Indeed, CCNE1 overexpression was associated with de novo resistance to CDK4/6 inhibitors in the PALOMA-3 trial, which assessed dual palbociclib and fulvestrant treatment in metastatic breast cancer (53), and with acquired resistance to CDK4/6 inhibitors in breast cancer cell lines in vitro (54). Few studies to date have explored the utility of CDK1/2 inhibition in breast cancer, either alone or combined with CDK4/6 (55, 56).
Intriguingly, genomic alterations in the mTOR and CDK/RB pathways, as well as the mTOR and focal adhesion pathways, preferentially co-occurred in metastases, whereby patients whose primary tumors exhibited genomic alterations in one of these pathways were more likely to give rise to metastases bearing genomic alterations in the other pathway. Furthermore, we found that alterations that are expected to affect the CDK/RB pathway, including gain of CCNE1 and loss of TP53, were significantly associated with mTOR activity in metastases, a finding that is consistent with accumulating evidence for extensive crosstalk between mTOR signaling and cell cycle regulators (57). Consequently, while studies of the ability of CDK4/6 inhibitors (PALLAS, monarchE) or mTOR inhibitors (SWOG/NRG/Alliance S1207) to prevent metastatic recurrence in early-stage breast cancer patients are underway, our findings that genomic alterations in these pathways co-occur in patients suggest that combined pharmacological inhibition of mTOR and CDKs may more effectively prevent, or treat, metastatic breast cancer than either agent alone. This prediction is supported by observations that dual inhibition of these targets results in more pronounced antitumor effects in breast cancer xenografts and cell lines in vitro (57).
The WNT signaling pathway is a highly conserved regulator of embryonic development (58) and has established roles in numerous aspects of cancer biology, including primary tumorigenesis, metastasis, and “stemness” (59, 60). Our study determined that patient metastases preferentially mutate the WNT pathway and hyperactivate canonical WNT signaling, as evidenced by an increase in both nuclear and membranous β-catenin. This hyperactivation is consistent with correlative histopathological studies demonstrating that β-catenin expression in primary breast cancers is associated with poor prognosis while being rarely localized to the nucleus (61). Despite these prognostic studies, surprisingly few studies have assessed WNT pathway activation in breast cancer metastases in patients. Whereas 2 studies failed to find nuclear expression of β-catenin in brain (62) or liver (63) metastases, 3 other studies assayed β-catenin in lymph node (64–66) and bone (65) metastases but did not address nuclear localization. Our study demonstrates that nuclear localization of β-catenin preferentially occurs in breast cancer metastases in patients and is evident in multiple organ sites (lymph node, liver, and brain), indicating that metastases broadly and preferentially activate the canonical WNT signaling pathway.
Interestingly, metastases exhibited increased expression of membranous β-catenin compared with primary tumors, despite the anticorrelative relationship between membrane and nuclear expression of β-catenin. An independent role for membranous β-catenin in promoting cellular adhesion (67) is consistent with our finding that the Focal Adhesion signaling pathway is preferentially mutated in metastases. In aggregate, our findings that metastases preferentially mutate the WNT Signaling pathway, exhibit canonical WNT activation, and overexpress membranous β-catenin, provide further evidence implicating WNT signaling in treatment-refractory, metastatic breast cancer.
Like the mTOR, CDK/RB, and WNT pathways, the cAMP/PKA signaling pathway was also preferentially mutated in metastases. Beyond our finding that cAMP/PKA pathway mutations are more common in patients exposed to either chemotherapy or radiation therapy, our study demonstrates that localization of activated PKA to the nucleus preferentially occurs in treatment-resistant metastases in patients, and that this occurs at multiple organ sites. Consistent with this, other studies have reported that phosphorylation of ER by PKA induces tamoxifen resistance in vitro (68) and that translocation of p-PKA to the nucleus mediates lung cancer metastasis in mice (69). Nevertheless, despite its ubiquitous presence and broad cellular effects, cellular responses to increased levels of cAMP are cell type and context specific (43, 70). It is therefore not surprising that conflicting evidence exists regarding the role of PKA in tumorigenesis, metastasis, and treatment resistance (71, 72). These apparent contradictions may be explained, at least in part, by the differential abundance of PKA regulatory subunits (73), subcellular localization of cAMP resulting from selective degradation by phosphodiesterases (PDEs) (74), and/or subcellular localization of activated PKA through binding to compartment-specific A-kinase anchoring proteins (75). Because both activators (e.g., forskolin) (76) and degraders (e.g., PDE4) (77) of cAMP are being evaluated in the clinic, the complex effects of cAMP/PKA pathway activation during cancer progression would benefit from further investigation.
Upstream of cAMP/PKA, we found preferential CN gain of the nongenomic PR, PAQR8, in metastases. This alteration was mutually exclusive with mutations in the canonical nuclear hormone receptors, ESR1 and PGR, and was associated with increased mTOR activity. Because the role of progesterone in breast cancer remains largely enigmatic, an exciting avenue of research will be to determine whether gain of PAQR8 results in increased mTOR or cAMP/PKA pathway activity, is an escape pathway from ER blockade, or represents a distinct mechanism of metastatic tumor dissemination or outgrowth distinct from canonical estrogen or progesterone signaling.
Within the metastasis-enriched Focal Adhesion pathway, CN gain of PTK6 would be anticipated to result in increased activity of its substrate, FAK (78). Three additional metastasis-enriched SMGs are predicted to influence this same pathway. MYLK promotes adhesion disassembly, cellular invasion, and formation of actin stress fibers (20, 79). PEAK1 promotes epithelial-mesenchymal transition via its association with the actin cytoskeleton and focal adhesions (80), and XIRP2 protects against depolymerization of actin filaments (81). Our observation that neither MYLK nor PEAK1 have been reported as SMGs in any other cancer type in TCGA is consistent with a specific role for the focal adhesion pathway in metastasis and/or treatment resistance that is distinct from its roles in primary tumorigenesis.
Though not an SMG in primary breast cancer, the metastasis-enriched SMG PALB2 is a known breast cancer susceptibility gene (82). PALB2 plays a critical role in homologous recombination through its interactions with BRCA1 and BRCA2 and, consistent with our findings here, has recently been shown to be recurrently mutated in metastatic breast cancer (6). Germline and/or somatic mutations in BRCA1, BRCA2, or PALB2 within metastases in our cohort were relatively frequent, were predicted to result in complete LOF via LOH or mutation of both alleles, and were associated with a concomitant increase in the number of LSTs, a marker of HRD. Unexpectedly, mutations in BRCA1, BRCA2, and PALB2 significantly co-occurred with mutations in HKMTs, which also were preferentially mutated in metastases, thereby suggesting a potential link between HRD and the disruption of epigenetic regulators during tumor progression. Together, our findings reveal that mutations affecting HRD-related genomic instability are selected for during tumor progression, suggesting that metastases in a large proportion of breast cancer patients may be rendered sensitive to platinum-based therapies and/or PARP inhibitors by mutations in these genes.
Several recent reports have provided valuable insights into the genomic landscape of metastatic breast cancer (4–12). However, the majority of these studies have been limited to targeted panels of genes previously implicated in cancer, unpaired tumor cohorts, small sample sizes, inclusion of treated primary tumors, and/or inference of CN based on non–genome-wide sequencing. The present study evaluates the genomic evolution of breast cancer both in a large cross section of metastases as well as in longitudinal paired primary-metastatic samples from the same patient. By expanding assays beyond targeted cancer gene panels, preferentially mutated and/or CN-altered genes could be identified in the metastatic setting beyond those frequently mutated in primary cancers. Moreover, preferentially mutated pathways were identified by leveraging the knowledge that concordant pathway alterations may result from mutations in multiple distinct pathway components, such that few individual genes are recurrently mutated. An additional unique feature of the current study is that multiple pathways identified as recurrently mutated at the genomic level were biochemically confirmed as altered in both paired and unpaired metastatic tumors.
Despite these strengths, limitations of the current study include the challenges of identifying subtype- and site-specific determinants of metastasis, which would require larger sample sizes, particularly given the high frequency of HR+ liver metastases in our cohort. Our study also did not address other known genomic drivers of cancer, such as noncoding variants, genomic translocations, and transcriptional and epigenetic dysregulation.
In aggregate, our study highlights the divergent genomic evolution of metastatic cancers, identifies potentially novel targets for combating metastatic progression and therapeutic escape, provides a genomic basis for the efficacy of mTOR, CDK4/6, and PARP inhibitors, and suggests WNT and nuclear PKA as drivers of breast cancer progression.
-
Methods
WES and sWGS were performed to detect somatic coding mutations and genome-wide CNAs in paired primary and metastatic tumors from 28 patients and in unpaired metastases from 38 additional patients. Tumors were available as formalin-fixed, paraffin-embedded (FFPE) blocks and/or OCT-embedded frozen core biopsies in the form of intact tumor blocks, intact core biopsies, or unstained 10-μm sections. ER, PR, and HER2 status were provided from clinical annotation and were determined using standard practices by IHC and/or FISH.
WES variants were called using an ensemble approach, combining calls from 3 standard pipelines. MutSigCV2 (19) was used to determine SMGs based on coding mutations and per-sample background mutation rates. SMG mutation frequencies were compared between metastases in this cohort and variants from TCGA-BRCA that were recalled using the same variant-calling pipeline. Filtering threshold and per-sample coverage were carefully considered when assessing increases in mutation frequencies.
sWGS CN values were estimated by adjusting read counts using QDNAseq (83) and were corrected by locus- and sample-specific parameters, tumor cellularity, and ploidy. GISTIC2 (27) was used to determine SARs based on genome-wide CN. Patient-oriented pathway analysis of somatic mutations was implemented based on approaches described in (39) and used pathway gene sets defined by KEGG (84).
p-S6, phospho-RB, β-catenin, and p-PKA were assessed using IHC to evaluate activity of the mTOR, CDK/RB, WNT, and cAMP/PKA pathways in 2 cohorts of primary and metastatic tumors, the majority of which were genomically assayed. One cohort consisted of paired tumors from the same patient; the other contained primary and metastatic tumors matched by tumor block age, preservation type, and the receptor subtype of the primary tumor from which the metastasis arose.
Three external data sets were used for in silico validation: recalled variants from primary tumors from TCGA-BRCA (3), reported CNAs from primary tumors in METABRIC (38), and reported variants from a cohort of metastatic tumors from Lefebvre et al. (6).
Clinical metadata, MutSigCV2 output, GISTIC2 output, and all described as well as relevant mutations are provided as Supplemental Tables. Raw WES and sWGS data are publicly accessible in the NCBI BioProject, PRJNA610817. For more information on study cohorts, data processing, and analyses presented in this study, see Supplemental Methods.
Statistics. Fisher’s exact test was used to compare mutant sample frequencies between primary and metastatic tumor cohorts (e.g., ESR1 mutation). The Wilcoxon rank-sum test was used to determine whether the number or extent of genomic events (e.g., CN values) within samples were significantly different between the 2 cohorts (or subsets within a cohort). The Wilcoxon signed-rank test was used to compare values between paired tumors. One-sided tests were used throughout where appropriate unless otherwise noted. Univariate linear regression was used to determine if 2 continuous features were correlated. Multiple testing correction and comparisons to permuted gene sets were used to evaluate significance, defined as FDR ≤ 0.10 and Pperm ≤ 0.10, and are specified where applicable.
Each SAR was tested to determine whether mean CN values were significantly different between paired primary and metastatic tumors (2-sided Wilcoxon’s signed-rank test) or whether the distributions of low-level CNA frequencies (percentage loss, percentage normal, and percentage gain) in primary tumors and the total metastatic tumor cohort were significantly different (χ2 test). Metastasis-enriched SMPs were identified by comparing the proportion of patients with private mutations solely in the primary tumor, solely in the metastatic tumor, or in both tumors using a McNemar test. Kaplan-Meier curve survival analyses were used to assess whether primary tumor mutations in SMGs were associated with decreased RFS in the TCGA-BRCA data set and whether primary tumor CNAs in implicated SARs were associated with decreased DSS in the METABRIC data set. The extent to which these associations were specific to implicated CN direction and were independent of CN burden was also assessed.
Study approval. Genomically assayed tumors were obtained from patients who provided informed consent to a research tumor biopsy as part of the METAMORPH Study (“Metastatic Markers of the Recurrent Phenotype”) and were ascertained from the clinical practices of the Rena Rowan Breast Center at the University of Pennsylvania, Philadelphia, Pennsylvania, USA (IRB 1, Protocol 818874). Additional tumor specimens were provided for IHC analyses by the TRACR (Breast Program Translational Cancer Resource) project (University of Pennsylvania School of Medicine IRB 1, Protocol 811475).
-
Author contributions
AD designed and implemented the TRACR project and the METAMORPH study with collaboration from the Center of Personalized Diagnostics (DL, JJDM), and oversaw the integration of clinical data with results of genomic analyses. SWS performed metastatic biopsies. DSE and CC provided clinical abstraction on patients. NNCS and MF evaluated archival tissue specimens and supervised their distribution and processing. YC and GKB processed tissue samples and supervised external and in-house sequencing. MRP designed sequencing projects in consultation with TCP and KNM. MRP designed and implemented bioinformatic analyses in consultation with TCP, DKP, and AS. DKP implemented variant calling of raw TCGA-BRCA sequencing data. TCP evaluated the role of sequencing coverage with mutational frequencies in TCGA-BRCA. NNCS supervised IHC antibody validation and execution. KLH facilitated generation and selection of representative IHC images. NGG coordinated the TRACR project which provided additional specimens for IHC. MRP and LAC designed and supervised all experiments, oversaw the interpretation of all analyses, and wrote the manuscript. All authors reviewed the manuscript.
-
Acknowledgments
We thank patients in the METAMORPH study and their families, without whom this study would not have been possible. We thank contributors at the University of Pennsylvania, including staff at the Tumor Tissue and Biospecimen Bank (TTAB) and the Pathology Clinical Service Center (PCSC). We thank Irfan Asangani and members of his laboratory for consultation on in-house high-throughput sequencing. We also thank Sara Chodosh for creating the graphical abstract for this manuscript. This study was supported by grants from the National Cancer Institute, the Emerson Collective Count Me In Challenge, the Breast Cancer Research Foundation, and FFANY/QVC “Shoes on Sale,” and by the Abramson Cancer Center, Penn Medicine and the 2-PREVENT Translational Center of Excellence.
Address correspondence to: Lewis A. Chodosh, Room 614 BRB II/III, 421 Curie Boulevard, Philadelphia, Pennsylvania 19104-6160, USA. Email: chodosh@pennmedicine.upenn.edu.
-
Footnotes
Conflict of interest: AD serves as principal investigator for clinical trials for which the University of Pennsylvania receives research funding from Novartis, Pfizer, Genentech, Calithera, and Menarini. JJDM received consultant fees from Novartis and Loxo Pharmaceuticals. SWS received consultant fees from Becton Dickinson and research funding from Cook Biotech and SillaJen. LAC received consultant fees for expert testimony on behalf of Imerys related to talc, and for consulting on behalf of Eli Lilly related to asbestos.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(8):4252–4265.https://doi.org/10.1172/JCI129941.
-
References
- Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends–an update. Cancer Epidemiol Biomarkers Prev. 2016;25(1):16–27.
- Pan H, et al. 20-Year risks of breast-cancer recurrence after stopping endocrine therapy at 5 years. N Engl J Med. 2017;377(19):1836–1846.
- Ciriello G, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163(2):506–519.
- Brastianos PK, et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;5(11):1164–1177.
- Bertucci F, et al. Comparative genomic analysis of primary tumors and metastases in breast cancer. Oncotarget. 2016;7(19):27208–27219.
- Lefebvre C, et al. Mutational profile of metastatic breast cancers: a retrospective analysis. PLoS Med. 2016;13(12):e1002201.
- Brown D, et al. Phylogenetic analysis of metastatic progression in breast cancer using somatic mutations and copy number aberrations. Nat Commun. 2017;8:14944. View this article via: PubMed Google Scholar
- Ng CKY, et al. Genetic heterogeneity in therapy-naïve synchronous primary breast cancers and their metastases. Clin Cancer Res. 2017;23(15):4402–4415.
- Robinson DR, et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548(7667):297–303.
- Yates LR, et al. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell. 2017;32(2):169–184.e7.
- Krøigård AB, et al. Identification of metastasis driver genes by massive parallel sequencing of successive steps of breast cancer progression. PLoS One. 2018;13(1):e0189887.
- Razavi P, et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell. 2018;34(3):427–438.e6.
- Maxwell KN, et al. Comparative clinical utility of tumor genomic testing and cell-free DNA in metastatic breast cancer. Breast Cancer Res Treat. 2017;164(3):627–638.
- Toy W, et al. Activating ESR1 mutations differentially affect the efficacy of ER antagonists. Cancer Discov. 2017;7(3):277–287.
- Watkins JA, Irshad S, Grigoriadis A, Tutt AN. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res. 2014;16(3):211.
- Maxwell KN, et al. BRCA locus-specific loss of heterozygosity in germline BRCA1 and BRCA2 carriers. Nat Commun. 2017;8(1):319.
- Gonzalez-Perez A, et al. IntOGen-mutations identifies cancer drivers across tumor types. Nat Methods. 2013;10(11):1081–1082.
- Xia B, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22(6):719–729.
- Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218.
- Webb DJ, et al. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol. 2004;6(2):154–161.
- Roy P, Perrin BJ. The stable actin core of mechanosensory stereocilia features continuous turnover of actin cross-linkers. Mol Biol Cell. 2018;29(15):1856–1865.
- Bristow JM, Reno TA, Jo M, Gonias SL, Klemke RL. Dynamic phosphorylation of tyrosine 665 in pseudopodium-enriched atypical kinase 1 (PEAK1) is essential for the regulation of cell migration and focal adhesion turnover. J Biol Chem. 2013;288(1):123–131.
- Sparling DP, Griesel BA, Weems J, Olson AL. GLUT4 enhancer factor (GEF) interacts with MEF2A and HDAC5 to regulate the GLUT4 promoter in adipocytes. J Biol Chem. 2008;283(12):7429–7437.
- Pineda B, et al. A two-gene epigenetic signature for the prediction of response to neoadjuvant chemotherapy in triple-negative breast cancer patients. Clin Epigenetics. 2019;11(1):33.
- Dorn KV, Hughes CE, Rohatgi R. A Smoothened-Evc2 complex transduces the Hedgehog signal at primary cilia. Dev Cell. 2012;23(4):823–835.
- Zhang H, et al. Elevated fibroblast growth factor signaling is critical for the pathogenesis of the dwarfism in Evc2/Limbin mutant mice. PLoS Genet. 2016;12(12):e1006510.
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41.
- Chen J, Lindblom A. Germline mutation screening of the STK11/LKB1 gene in familial breast cancer with LOH on 19p. Clin Genet. 2000;57(5):394–397.View this article via: PubMed Google Scholar
- Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9(8):563–575.
- Li J, et al. Loss of LKB1 disrupts breast epithelial cell polarity and promotes breast cancer metastasis and invasion. J Exp Clin Cancer Res. 2014;33:70. View this article via: PubMed Google Scholar
- Cancer Genome Atlas Network . Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70.
- Karteris E, et al. Progesterone signaling in human myometrium through two novel membrane G protein-coupled receptors: potential role in functional progesterone withdrawal at term. Mol Endocrinol. 2006;20(7):1519–1534.
- Zhou L, et al. Progesterone suppresses triple-negative breast cancer growth and metastasis to the brain via membrane progesterone receptor α. Int J Mol Med. 2017;40(3):755–761.
- Brauer PM, Tyner AL. Building a better understanding of the intracellular tyrosine kinase PTK6 - BRK by BRK. Biochim Biophys Acta. 2010;1806(1):66–73.View this article via: PubMed Google Scholar
- Ito K, Park SH, Nayak A, Byerly JH, Irie HY. PTK6 inhibition suppresses metastases of triple-negative breast cancer via SNAIL-dependent E-cadherin regulation. Cancer Res. 2016;76(15):4406–4417.
- Ito K, et al. PTK6 regulates growth and survival of endocrine therapy-resistant ER+ breast cancer cells. NPJ Breast Cancer. 2017;3:45. View this article via: PubMed Google Scholar
- Harvey AJ, Crompton MR. Use of RNA interference to validate Brk as a novel therapeutic target in breast cancer: Brk promotes breast carcinoma cell proliferation. Oncogene. 2003;22(32):5006–5010.
- Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352.
- Boca SM, Kinzler KW, Velculescu VE, Vogelstein B, Parmigiani G. Patient-oriented gene set analysis for cancer mutation data. Genome Biol. 2010;11(11):R112.
- Masui K, et al. Glucose-dependent acetylation of Rictor promotes targeted cancer therapy resistance. Proc Natl Acad Sci U S A. 2015;112(30):9406–9411.
- Harter PN, et al. Immunohistochemical assessment of phosphorylated mTORC1-pathway proteins in human brain tumors. PLoS One. 2015;10(5):e0127123.
- Kannengiesser C, et al. Functional, structural, and genetic evaluation of 20 CDKN2A germ line mutations identified in melanoma-prone families or patients. Hum Mutat. 2009;30(4):564–574.
- Chin KV, et al. Reinventing the wheel of cyclic AMP: novel mechanisms of cAMP signaling. Ann N Y Acad Sci. 2002;968:49–64.
- Zhao JH, Reiske H, Guan JL. Regulation of the cell cycle by focal adhesion kinase. J Cell Biol. 1998;143(7):1997–2008.
- Lee FY, et al. The mTOR-FAK mechanotransduction signaling axis for focal adhesion maturation and cell proliferation. Am J Transl Res. 2017;9(4):1603–1617.View this article via: PubMed Google Scholar
- Wang Q, et al. PI3K-p110α mediates resistance to HER2-targeted therapy in HER2+, PTEN-deficient breast cancers. Oncogene. 2016;35(27):3607–3612.
- Beelen K, et al. PI3K/AKT/mTOR pathway activation in primary and corresponding metastatic breast tumors after adjuvant endocrine therapy. Int J Cancer. 2014;135(5):1257–1263.
- Royce ME, Osman D. Everolimus in the treatment of metastatic breast cancer. Breast Cancer (Auckl). 2015;9:73–79.View this article via: PubMed Google Scholar
- Pierobon M, et al. Enrichment of PI3K-AKT-mTOR pathway activation in hepatic metastases from breast cancer. Clin Cancer Res. 2017;23(16):4919–4928.
- Vasan N, Toska E, Scaltriti M. Overview of the relevance of PI3K pathway in HR-positive breast cancer. Ann Oncol. 2019;30 Suppl 10:x3–x11.
- Miller TW, et al. ERα-dependent E2F transcription can mediate resistance to estrogen deprivation in human breast cancer. Cancer Discov. 2011;1(4):338–351.
- de Groot AF, Kuijpers CJ, Kroep JR. CDK4/6 inhibition in early and metastatic breast cancer: A review. Cancer Treat Rev. 2017;60:130–138.
- Turner NC, et al. Abstract CT039: Cyclin E1 (CCNE1) expression associates with benefit from palbociclib in metastatic breast cancer (MBC) in the PALOMA3 trial. Cancer Res. 2018;78(13 supplement):CT039. View this article via: CrossRef Google Scholar
- Yang C, et al. Acquired CDK6 amplification promotes breast cancer resistance to CDK4/6 inhibitors and loss of ER signaling and dependence. Oncogene. 2017;36(16):2255–2264.
- Patel P, Tsiperson V, Gottesman SRS, Somma J, Blain SW. Dual inhibition of CDK4 and CDK2 via targeting p27 tyrosine phosphorylation induces a potent and durable response in breast cancer cells. Mol Cancer Res. 2018;16(3):361–377.
- Xia Q, Cai Y, Peng R, Wu G, Shi Y, Jiang W. The CDK1 inhibitor RO3306 improves the response of BRCA-proficient breast cancer cells to PARP inhibition. Int J Oncol. 2014;44(3):735–744.
- Michaloglou C, et al. Combined inhibition of mTOR and CDK4/6 is required for optimal blockade of E2F function and long-term growth inhibition in estrogen receptor-positive breast cancer. Mol Cancer Ther. 2018;17(5):908–920.
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810.
- Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36(11):1461–1473.
- Gunther EJ, et al. Impact of p53 loss on reversal and recurrence of conditional Wnt-induced tumorigenesis. Genes Dev. 2003;17(4):488–501.
- Borcherding N, et al. Re-evaluating E-cadherin and β-catenin: a pan-cancer proteomic approach with an emphasis on breast cancer. Am J Pathol. 2018;188(8):1910–1920.
- Klemm F, et al. β-catenin-independent WNT signaling in basal-like breast cancer and brain metastasis. Carcinogenesis. 2011;32(3):434–442.
- Bleckmann A, et al. β-catenin-independent WNT signaling and Ki67 in contrast to the estrogen receptor status are prognostic and associated with poor prognosis in breast cancer liver metastases. Clin Exp Metastasis. 2016;33(4):309–323.
- Bukholm IK, Nesland JM, Børresen-Dale AL. Re-expression of E-cadherin, alpha-catenin and beta-catenin, but not of gamma-catenin, in metastatic tissue from breast cancer patients. J Pathol. 2000;190(1):15–19.
- Kasoha M, Bohle RM, Seibold A, Gerlinger C, Juhasz-Böss I, Solomayer EF. Dickkopf-1 (Dkk1) protein expression in breast cancer with special reference to bone metastases. Clin Exp Metastasis. 2018;35(8):763–775.
- Khramtsov AI, Khramtsova GF, Tretiakova M, Huo D, Olopade OI, Goss KH. Wnt/beta-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am J Pathol. 2010;176(6):2911–2920.
- Peifer M, McCrea PD, Green KJ, Wieschaus E, Gumbiner BM. The vertebrate adhesive junction proteins beta-catenin and plakoglobin and the Drosophila segment polarity gene armadillo form a multigene family with similar properties. J Cell Biol. 1992;118(3):681–691.
- Michalides R, et al. Tamoxifen resistance by a conformational arrest of the estrogen receptor alpha after PKA activation in breast cancer. Cancer Cell. 2004;5(6):597–605.
- Zuo Z, et al. High mobility group Box-1 inhibits cancer cell motility and metastasis by suppressing activation of transcription factor CREB and nWASP expression. Oncotarget. 2014;5(17):7458–7470.
- Stork PJ, Schmitt JM. Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol. 2002;12(6):258–266.
- Pattabiraman DR, et al. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science. 2016;351(6277):aad3680.
- Xu S, Zhou W, Ge J, Zhang Z. Prostaglandin E2 receptor EP4 is involved in the cell growth and invasion of prostate cancer via the cAMP‑PKA/PI3K‑Akt signaling pathway. Mol Med Rep. 2018;17(3):4702–4712.View this article via: PubMed Google Scholar
- Bossis I, Stratakis CA. Minireview: PRKAR1A: normal and abnormal functions. Endocrinology. 2004;145(12):5452–5458.
- Sample V, DiPilato LM, Yang JH, Ni Q, Saucerman JJ, Zhang J. Regulation of nuclear PKA revealed by spatiotemporal manipulation of cyclic AMP. Nat Chem Biol. 2012;8(4):375–382.
- Carnegie GK, Scott JD. A-kinase anchoring proteins and neuronal signaling mechanisms. Genes Dev. 2003;17(13):1557–1568.
- Sapio L, et al. The natural cAMP elevating compound forskolin in cancer therapy: Is it time? J Cell Physiol. 2017;232(5):922–927.
- Omar F, et al. Small-molecule allosteric activators of PDE4 long form cyclic AMP phosphodiesterases. Proc Natl Acad Sci U S A. 2019;116(27):13320–13329.
- Zheng Y, Gierut J, Wang Z, Miao J, Asara JM, Tyner AL. Protein tyrosine kinase 6 protects cells from anoikis by directly phosphorylating focal adhesion kinase and activating AKT. Oncogene. 2013;32(36):4304–4312.
- Hsu YC, et al. Genome-wide analysis of three-way interplay among gene expression, cancer cell invasion and anti-cancer compound sensitivity. BMC Med. 2013;11:106. View this article via: PubMed Google Scholar
- Agajanian M, et al. PEAK1 Acts as a molecular switch to regulate context-dependent TGFβ responses in breast cancer. PLoS One. 2015;10(8):e0135748.
- Pacholsky D, et al. Xin repeats define a novel actin-binding motif. J Cell Sci. 2004;117(pt 22):5257–5268.View this article via: PubMed Google Scholar
- Couch FJ, et al. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol. 2017;3(9):1190–1196.
- Scheinin I, et al. DNA copy number analysis offresh and formalin-fixed specimens by shallowwhole-genome sequencing with identificationand exclusion of problematic regions in thegenome assembly. Genome Res. 2014;24(12):2022–2032.
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361.
-
Version history
- Version 1 (July 13, 2020): Electronic publication
- Version 2 (August 3, 2020): Print issue publication



Copyright © 2026 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)