Advertisement
Concise CommunicationEndocrinology Free access | 10.1172/JCI126309
β Cell–intrinsic β-arrestin 1 signaling enhances sulfonylurea-induced insulin secretion
Luiz F. Barella,1 Mario Rossi,1 Lu Zhu,1 Yinghong Cui,1 Fang C. Mei,2 Xiaodong Cheng,2 Wei Chen,3 Vsevolod V. Gurevich,4 and Jürgen Wess1
1Molecular Signaling Section, Laboratory of Bioorganic Chemistry, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, Maryland, USA.
2Department of Integrative Biology & Pharmacology, University of Texas Health Science Center at Houston, Houston, Texas, USA.
3Department of Medicine, Duke University Medical Center, Durham, North Carolina, USA.
4Department of Pharmacology, Vanderbilt University, Nashville, Tennessee, USA.
Address correspondence to: Jürgen Wess, Laboratory of Bioorganic Chemistry, Molecular Signaling Section, National Institute of Diabetes and Digestive and Kidney Diseases, Bldg. 8A, Room B1A-05, 8 Center Drive, Bethesda, Maryland 20892, USA. Phone: 301.402.3589; E-mail: jurgenw@niddk.nih.gov.
Find articles by
Barella, L.
in:
PubMed
|
Google Scholar
|

1Molecular Signaling Section, Laboratory of Bioorganic Chemistry, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, Maryland, USA.
2Department of Integrative Biology & Pharmacology, University of Texas Health Science Center at Houston, Houston, Texas, USA.
3Department of Medicine, Duke University Medical Center, Durham, North Carolina, USA.
4Department of Pharmacology, Vanderbilt University, Nashville, Tennessee, USA.
Address correspondence to: Jürgen Wess, Laboratory of Bioorganic Chemistry, Molecular Signaling Section, National Institute of Diabetes and Digestive and Kidney Diseases, Bldg. 8A, Room B1A-05, 8 Center Drive, Bethesda, Maryland 20892, USA. Phone: 301.402.3589; E-mail: jurgenw@niddk.nih.gov.
Find articles by
Rossi, M.
in:
PubMed
|
Google Scholar
|

1Molecular Signaling Section, Laboratory of Bioorganic Chemistry, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, Maryland, USA.
2Department of Integrative Biology & Pharmacology, University of Texas Health Science Center at Houston, Houston, Texas, USA.
3Department of Medicine, Duke University Medical Center, Durham, North Carolina, USA.
4Department of Pharmacology, Vanderbilt University, Nashville, Tennessee, USA.
Address correspondence to: Jürgen Wess, Laboratory of Bioorganic Chemistry, Molecular Signaling Section, National Institute of Diabetes and Digestive and Kidney Diseases, Bldg. 8A, Room B1A-05, 8 Center Drive, Bethesda, Maryland 20892, USA. Phone: 301.402.3589; E-mail: jurgenw@niddk.nih.gov.
Find articles by
Zhu, L.
in:
PubMed
|
Google Scholar
|

1Molecular Signaling Section, Laboratory of Bioorganic Chemistry, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, Maryland, USA.
2Department of Integrative Biology & Pharmacology, University of Texas Health Science Center at Houston, Houston, Texas, USA.
3Department of Medicine, Duke University Medical Center, Durham, North Carolina, USA.
4Department of Pharmacology, Vanderbilt University, Nashville, Tennessee, USA.
Address correspondence to: Jürgen Wess, Laboratory of Bioorganic Chemistry, Molecular Signaling Section, National Institute of Diabetes and Digestive and Kidney Diseases, Bldg. 8A, Room B1A-05, 8 Center Drive, Bethesda, Maryland 20892, USA. Phone: 301.402.3589; E-mail: jurgenw@niddk.nih.gov.
Find articles by Cui, Y. in: PubMed | Google Scholar
1Molecular Signaling Section, Laboratory of Bioorganic Chemistry, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, Maryland, USA.
2Department of Integrative Biology & Pharmacology, University of Texas Health Science Center at Houston, Houston, Texas, USA.
3Department of Medicine, Duke University Medical Center, Durham, North Carolina, USA.
4Department of Pharmacology, Vanderbilt University, Nashville, Tennessee, USA.
Address correspondence to: Jürgen Wess, Laboratory of Bioorganic Chemistry, Molecular Signaling Section, National Institute of Diabetes and Digestive and Kidney Diseases, Bldg. 8A, Room B1A-05, 8 Center Drive, Bethesda, Maryland 20892, USA. Phone: 301.402.3589; E-mail: jurgenw@niddk.nih.gov.
Find articles by Mei, F. in: PubMed | Google Scholar
1Molecular Signaling Section, Laboratory of Bioorganic Chemistry, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, Maryland, USA.
2Department of Integrative Biology & Pharmacology, University of Texas Health Science Center at Houston, Houston, Texas, USA.
3Department of Medicine, Duke University Medical Center, Durham, North Carolina, USA.
4Department of Pharmacology, Vanderbilt University, Nashville, Tennessee, USA.
Address correspondence to: Jürgen Wess, Laboratory of Bioorganic Chemistry, Molecular Signaling Section, National Institute of Diabetes and Digestive and Kidney Diseases, Bldg. 8A, Room B1A-05, 8 Center Drive, Bethesda, Maryland 20892, USA. Phone: 301.402.3589; E-mail: jurgenw@niddk.nih.gov.
Find articles by
Cheng, X.
in:
PubMed
|
Google Scholar
|

1Molecular Signaling Section, Laboratory of Bioorganic Chemistry, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, Maryland, USA.
2Department of Integrative Biology & Pharmacology, University of Texas Health Science Center at Houston, Houston, Texas, USA.
3Department of Medicine, Duke University Medical Center, Durham, North Carolina, USA.
4Department of Pharmacology, Vanderbilt University, Nashville, Tennessee, USA.
Address correspondence to: Jürgen Wess, Laboratory of Bioorganic Chemistry, Molecular Signaling Section, National Institute of Diabetes and Digestive and Kidney Diseases, Bldg. 8A, Room B1A-05, 8 Center Drive, Bethesda, Maryland 20892, USA. Phone: 301.402.3589; E-mail: jurgenw@niddk.nih.gov.
Find articles by Chen, W. in: PubMed | Google Scholar
1Molecular Signaling Section, Laboratory of Bioorganic Chemistry, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, Maryland, USA.
2Department of Integrative Biology & Pharmacology, University of Texas Health Science Center at Houston, Houston, Texas, USA.
3Department of Medicine, Duke University Medical Center, Durham, North Carolina, USA.
4Department of Pharmacology, Vanderbilt University, Nashville, Tennessee, USA.
Address correspondence to: Jürgen Wess, Laboratory of Bioorganic Chemistry, Molecular Signaling Section, National Institute of Diabetes and Digestive and Kidney Diseases, Bldg. 8A, Room B1A-05, 8 Center Drive, Bethesda, Maryland 20892, USA. Phone: 301.402.3589; E-mail: jurgenw@niddk.nih.gov.
Find articles by
Gurevich, V.
in:
PubMed
|
Google Scholar
|

1Molecular Signaling Section, Laboratory of Bioorganic Chemistry, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, Maryland, USA.
2Department of Integrative Biology & Pharmacology, University of Texas Health Science Center at Houston, Houston, Texas, USA.
3Department of Medicine, Duke University Medical Center, Durham, North Carolina, USA.
4Department of Pharmacology, Vanderbilt University, Nashville, Tennessee, USA.
Address correspondence to: Jürgen Wess, Laboratory of Bioorganic Chemistry, Molecular Signaling Section, National Institute of Diabetes and Digestive and Kidney Diseases, Bldg. 8A, Room B1A-05, 8 Center Drive, Bethesda, Maryland 20892, USA. Phone: 301.402.3589; E-mail: jurgenw@niddk.nih.gov.
Find articles by Wess, J. in: PubMed | Google Scholar
Published June 11, 2019 - More info
J Clin Invest. 2019;129(9):3732–3737. https://doi.org/10.1172/JCI126309.
© 2019 American Society for Clinical Investigation
Received: November 19, 2018; Accepted: June 4, 2019
-
Abstract
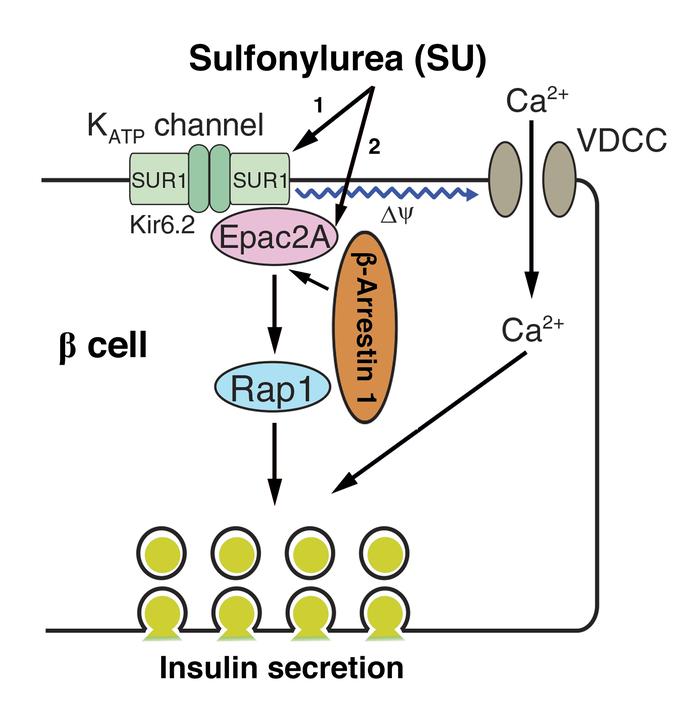
β-Arrestin 1 and 2 (Barr1 and Barr2, respectively) are intracellular signaling molecules that regulate many important metabolic functions. We previously demonstrated that mice lacking Barr2 selectively in pancreatic β cells showed pronounced metabolic impairments. Here we investigated whether Barr1 plays a similar role in regulating β cell function and whole-body glucose homeostasis. Initially, we inactivated the Barr1 gene in β cells of adult mice (β-barr1-KO mice). β-barr1-KO mice did not display any obvious phenotypes in a series of in vivo and in vitro metabolic tests. However, glibenclamide and tolbutamide, 2 widely used antidiabetic drugs of the sulfonylurea (SU) family, showed greatly reduced efficacy in stimulating insulin secretion in the KO mice in vivo and in perifused KO islets in vitro. Additional in vivo and in vitro studies demonstrated that Barr1 enhanced SU-stimulated insulin secretion by promoting SU-mediated activation of Epac2. Pull-down and coimmunoprecipitation experiments showed that Barr1 can directly interact with Epac2 and that SUs such as glibenclamide promote Barr1/Epac2 complex formation, triggering enhanced Rap1 signaling and insulin secretion. These findings suggest that strategies aimed at promoting Barr1 signaling in β cells may prove useful for the development of efficacious antidiabetic drugs.
-
Introduction
Accumulating evidence suggests that β-arrestins play key roles in regulating many important metabolic functions including β cell activity (1). The 2 β-arrestin isoforms (β-arrestin 1 and 2; referred to as Barr1 and Barr2 herein; also known as arrestin 2 and 3, respectively) play key roles in the desensitization and internalization of nearly all G protein–coupled receptors (GPCRs) (2). In addition, many studies suggest that β-arrestins can also act as signaling molecules in their own right (3, 4).
We recently demonstrated that β cell Barr2 is essential for the proper function of pancreatic β cells (5). In contrast, the potential role of Barr1 in regulating β cell activity and insulin secretion remains largely unexplored. To address this issue, we selectively deleted the Barr1 gene in pancreatic β cells of adult mice and subjected the resulting mutant animals to a series of metabolic tests.
Sulfonylurea drugs (SUs) have been a cornerstone for the therapy of type 2 diabetes (T2D) for more than 50 years (6). We demonstrated that β cell Barr1 deficiency selectively impairs SU-induced insulin release in vivo and in vitro. We also found that β cell Barr1 can exist in a complex with Epac2 and that this interaction promotes Epac2 activity. Our data are consistent with the concept that Barr1 plays an important role in regulating SU-dependent Epac2/Rap1 signaling in β cells, leading to enhanced SU-induced insulin secretion. These findings suggest that agents that can enhance Barr1 signaling in β cells may prove useful as efficacious antidiabetic drugs.
-
Results and Discussion
Selective deletion of the Barr1 gene in β cells of adult mice. We employed a conditional gene deletion strategy to selectively inactivate the Barr1 gene in β cells of adult mice. Several studies have shown that tamoxifen (TMX) induces Cre activity in Pdx1-Cre-ERTM transgenic mice selectively in pancreatic β cells (7, 8). We crossed Pdx1-Cre-ERTM mice with homozygous floxed Barr1 mice in which exon 2 was flanked by loxP sites (fl/fl Barr1 mice) (9). Subsequent matings led to the generation of fl/fl Barr1–Pdx1-Cre-ERTM mice and fl/fl Barr1 control littermates, which served as control animals throughout this study. All mouse lines used were maintained on a C57BL/6 background.
We injected fl/fl Barr1–Pdx1-Cre-ERTM mice and their control littermates (8-week-old males) for 6 consecutive days with TMX (2 mg i.p. per mouse per day) to induce Cre activity and Barr1 inactivation selectively in pancreatic β cells (7, 8). Gene expression and Western blotting studies confirmed the selective deletion of Barr1 in pancreatic islets of TMX-injected fl/fl Barr1–Pdx1-Cre-ERTM mice (Supplemental Figure 1A, C). Herein, we refer to the TMX-treated fl/fl Barr1–Pdx1-Cre-ERTM mice simply as β-barr1-KO mice. Barr2 expression levels remained unaffected by the Barr1 deletion in islets and other tissues from β-barr1-KO mice (Supplemental Figure 1B).
Untreated β-barr1-KO mice do not show any obvious metabolic deficits. β-barr1-KO mice and their control littermates did not show any statistically significant differences in body weight or fed and fasting blood glucose and plasma insulin levels (Supplemental Figure 2). Likewise, both groups of mice displayed similar blood glucose excursions in i.p. glucose and insulin tolerance tests, and showed similar increases in plasma insulin levels following injection of a glucose bolus (2 g/kg i.p.) (Supplemental Figure 3). Moreover, perifusion of islets from both control and β-barr1-KO mice with a high concentration of glucose (16 mM) triggered comparable insulin responses (Supplemental Figure 4).
Islet morphometric studies showed that β cell mass and islet size were unaltered by β cell Barr1 deficiency (Supplemental Figure 5). Additionally, we did not detect any significant changes in the expression levels of key genes and proteins involved in β cell function and maintenance (Supplemental Figure 6 and Supplemental Figure 7).
Enhanced insulin secretion caused by β cell GPCR signaling remains unaffected by β cell Barr1 deficiency. Insulin secretion from pancreatic β cells is stimulated by the activity of various GPCRs, including β cell M3 muscarinic (10, 11) and β cell GLP-1 receptors (12, 13). To test whether β cell Barr1 deficiency affected M3 and GLP-1 receptor–mediated insulin release in vivo, we injected control and β-barr1-KO mice with bethanechol (2 mg/kg i.p.), a muscarinic receptor agonist, or exendin-4 (12 nmol/kg i.p.), a GLP-1 receptor agonist. Previous studies have shown that treatment of mice with these 2 agonists leads to marked increases in plasma insulin levels that require the presence of β cell M3 (14) or GLP-1 receptors (15, 16), respectively. Both control and mutant mice exhibited similar insulin responses when treated with either bethanechol or exendin-4 alone or in combination with a glucose bolus (2 g/kg i.p.) (Supplemental Figure 8). In agreement with these in vivo data, islet perifusion studies carried out in the presence of 16 mM glucose demonstrated that acetylcholine (0.5 μM), the endogenous β cell M3 receptor agonist, and GLP-1 (0.1 μM) caused similar increases in GSIS in control and mutant islets (Supplemental Figure 9). In addition, we showed that GLP-1–stimulated insulin secretion is not impaired in islets derived from whole-body Barr1-KO mice (Supplemental Figure 10).
Taken together, these data strongly suggest that the ability of M3 and GLP-1 receptors to promote insulin release does not require the presence of Barr1 in mouse islets, in contrast to a previous finding that Barr1 is essential for GLP-1 receptor–dependent enhancement of insulin secretion in rat INS-1 insulinoma cells (17).
Deletion of Barr1 in β cells greatly reduces SU-induced insulin secretion in vivo. SUs enhance insulin secretion by binding to the SUR1 subunit of the K+ATP channel expressed by pancreatic β cells, leading to K+ATP channel closure, membrane depolarization, and subsequent insulin release (18). However, SUs can also promote insulin secretion via binding to and activation of β cell Epac2 (19–22), a cAMP binding protein that possesses guanine nucleotide exchange factor (GEF) activity toward Rap1 (23). Epac2/Rap1 signaling is known to play a key role in trafficking insulin granules to the plasma membrane (23). In a previous study, Mangmool et al. (24) demonstrated that Epac1, a close structural homolog of Epac2, can interact with β-arrestins in cardiac tissue and cultured cells, thereby modulating cellular signaling. On the basis of these findings, we explored the possibility that the lack of β cell Barr1 may affect SU-induced insulin section.
To test this hypothesis, we initially injected control and β-barr1-KO mice with glibenclamide (5 mg/kg i.p.) or tolbutamide (25 mg/kg i.p.), 2 commonly used SU drugs. We found that glibenclamide- and tolbutamide-stimulated increases in plasma insulin levels were significantly reduced in β-barr1-KO mice (Figure 1, A and B). In contrast, this deficit was not observed after treatment of control and mutant mice with gliclazide (10 mg/kg i.p.), another SU drug (Figure 1C). Previous work has shown that glibenclamide and tolbutamide, but not gliclazide, can activate β cell Epac2 (in addition to inhibiting K+ATP channels), thus contributing to SU-induced insulin secretion (19–21). Islet Sur1 and Epac2 expression remained unaffected by β cell Barr1 deficiency (Supplemental Figure 6 and Supplemental Figure 7). Thus, our observations strongly suggest that Barr1 plays an important role in regulating SU-dependent Epac2/Rap1 signaling in β cells.
 Figure 1
Figure 1Glibenclamide- and tolbutamide-stimulated insulin secretion is greatly impaired in β-barr1-KO mice. (A–C) Freely fed mice were injected i.p. with glibenclamide (5 mg/kg) (A), tolbutamide (25 mg/kg) (B), or gliclazide (10 mg/kg) (C). Plasma insulin levels were measured at the indicated time points using blood collected from the tail vein. All experiments were carried out with male littermates that were 10 to 12 weeks old. Actual basal plasma insulin levels were (in ng/mL): (A) Control: 1.33 ± 0.29, KO: 1.58 ± 0.25; (B) Control: 1.49 ± 0.14, KO: 1.64 ± 0.29; (C) Control: 1.38 ± 0.13, KO: 1.25 ± 0.15. Data are presented as mean ± SEM (n = 8 animals/group). *P < 0.05; **P < 0.01 (2-way ANOVA followed by Tukey’s post hoc test).
Studies with perifused islets. To confirm that the reduced efficacy of glibenclamide and tolbutamide to stimulate insulin secretion in β-barr1-KO mice in vivo was indeed caused by signaling deficits in pancreatic β cells, we performed a series of islet perifusion studies. In these experiments, glibenclamide (10 nM) and tolbutamide (500 μM) caused significantly smaller insulin responses in β-barr1-KO islets as compared with control islets (Figure 2, A and B). In contrast, gliclazide (10 μM) was able to stimulate insulin secretion to a similar extent in control and mutant islets (Figure 2C). The insulin content of control and KO islets did not differ significantly from each other (control: 532 ± 57 ng/mL; KO: 392 ± 54 ng/mL; 5–6 batches of 10 islets each, prepared from 3 different mice per genotype).
 Figure 2
Figure 2The absence of β cell Barr1 in isolated islets reduces insulin secretion in response to glibenclamide and tolbutamide, but not gliclazide. (A–C) Islets from control and β-barr1-KO mice were perifused with 3 mM glucose, either in the presence or absence of glibenclamide (10 nM) (A), tolbutamide (500 μM) (B), or gliclazide (10 μM) (C). The amount of secreted insulin was normalized to DNA content. All islets were prepared from male littermates that were 12 to 15 weeks old. Data are mean ± SEM (5 or 6 perifusions with 50 islets per perifusion chamber; islets were isolated from 6 mice per genotype). *P < 0.05 (2-tailed Student’s t test).
To further test the hypothesis that Barr1 is required for SU-mediated Epac2 activation, we performed additional islet perifusion studies using a specific Epac2 inhibitor, ESI-05 (25). Glibenclamide-induced insulin secretion from control islets was greatly decreased in the presence of ESI-05 (10 μM) (Figure 3A), consistent with the predicted role of Epac2 in contributing to glibenclamide-stimulated insulin secretion (19–21). In contrast, ESI-05 treatment had no significant effect on glibenclamide-stimulated insulin release from β-barr1-KO islets (Figure 3B). Likewise, ESI-05 had no significant effect on gliclazide-induced secretion from WT islets (Supplemental Figure 11). These data further support the notion that Barr1 plays a critical role in promoting the stimulation of Epac2 by SUs in β cells.
 Figure 3
Figure 3Control islets, but not β-barr1-KO islets, show greatly reduced glibenclamide-induced insulin secretion in the presence of a selective Epac2 inhibitor (ESI-05). (A, B) Islets from control (A) and β-barr1-KO mice (B) were perifused with 3 mM glucose, either in the presence or absence of 10 nM glibenclamide (GLB) or a mixture of 10 nM GLB and ESI-05 (10 μM), a selective Epac2 inhibitor. All experiments were carried out with male littermates that were 12 to 15 weeks old. Data are mean ± SEM (5 or 6 perifusions with 50 islets per perifusion chamber; islets were isolated from 6 mice per genotype; 2-tailed Student’s t test).
To exclude the possibility that Epac2 function was generally impaired in β-barr1-KO islets, we stimulated β-barr1-KO and control islets with 8-pCPT-2-O-Me-cAMP-AM (8-pCPT), an Epac-specific agonist. 8-pCPT (5 μM) treatment resulted in comparable increases in insulin secretion in mutant and control islets, suggesting that cAMP-dependent Epac2 activation and the downstream signaling pathway triggering insulin secretion remain intact in β cells lacking Barr1 (Supplemental Figure 12).
To probe the potential role of Barr2 in SU-induced insulin secretion, we studied perifused islets prepared from whole-body Barr2-KO mice. We found that both glibenclamide- and gliclazide-induced stimulation of insulin secretion was significantly impaired in the Barr2 mutant islets, as compared with WT control islets (Supplemental Figure 13). Since gliclazide does not require Epac2 for efficient insulin secretion and β cell Barr2 deficiency causes greatly reduced glucose- and KCl-induced insulin secretion (5), the decreased activity of SUs in Barr2-deficient islets is most likely due to the generalized secretory deficit displayed by the Barr2 mutant islets (5).
Barr1 directly interacts with Epac2. We next examined whether Barr1 was able to directly interact with Epac2. We performed pull-down assays using purified Barr1 protein and a purified GST-Epac2 fusion protein. The GST-Epac2 fusion protein (5 μg) or GST alone (negative control; 5 μg) were immobilized to a glutathione affinity resin. The immobilized proteins were then incubated with purified Barr1 (5 μg) for 1 hour at 4°C, followed by thorough washing. Bound proteins were then eluted with glutathione-containing buffer. Eluates were analyzed by SDS-PAGE/Western blotting using an anti-Barr1 antibody. This analysis showed that Barr1 protein was able to interreact with Epac2 in a specific fashion (Supplemental Figure 14). The addition of glibenclamide (100 nM), 8-pCPT (1 μM), or a mixture of glibenclamide (100 nM) and 8-pCPT (1 μM) had no significant effect on the intensity of the Barr1 immunoreactive bands.
Coimmunoprecipitation of a Barr1/Epac2 complex in a mouse β cell line. We next performed coimmunoprecipitation assays using MIN6-K8 mouse insulinoma cells (26) overexpressing Barr1 and a FLAG-tagged version of Epac2. Overexpression of the 2 proteins was achieved by the use of recombinant adenoviruses. The engineered MIN6-K8 cells were then incubated with glibenclamide (1 μM) for 30 or 60 minutes. Subsequently, cell lysates were subjected to immunoprecipitation with either an anti-FLAG antibody or rabbit IgG (negative control). Immunoprecipitated proteins were probed with an anti-Barr1 antibody by Western blotting. Using this strategy, Barr1 protein (~50 kDa) could only be detected in immunoprecipitates exposed to the anti-FLAG antibody (Figure 4A). Importantly, glibenclamide promoted the Barr1/Epac2 interaction in a time-dependent fashion (Figure 4, A and B). These data support the idea that Barr1 can exist in a complex with Epac2 and that SUs such as glibenclamide are able to further stabilize this complex. Since glibenclamide did not promote Barr1/Epac2 binding in the pull-down assay (see previous paragraph), SU-stimulated Barr1 binding to Epac2 appears to require additional proteins/factors that are only present in vivo.
 Figure 4
Figure 4Glibenclamide promotes the interaction of Barr1 with Epac2 and stimulates Rap1 activation in a Barr1-dependent fashion. (A) Coimmunoprecipitation was performed with MIN6-K8 cells infected with adenoviruses encoding Barr1 and Epac2-FLAG. Cells were stimulated with 1 μM glibenclamide (GLB) for 30 or 60 minutes. Cell lysates were incubated with an anti-FLAG antibody or rabbit IgG (negative control), and immunoprecipitated proteins were probed with an anti-Barr1 antibody by Western blotting. Data from a representative experiment are shown. (B) Quantification of the amount of Barr1 detected by Western blotting in the coimmunoprecipitation studies shown in A. Data are mean ± SEM of 3 independent experiments. (C) Efficient knockdown of Barr1 gene expression in MIN6-K8 cells by the use of Barr1 siRNA (n = 4). Con, scrambled control siRNA. (D) GLB treatment promotes the formation of Rap1-GTP in a Barr1-dependent fashion in MIN6-K8 cells. MIN6-K8 cells treated with scrambled control or Barr1 siRNA were incubated with GLB (100 nM) for 30 minutes and Rap1-GTP and total Rap1 levels were determined by Western blotting. Representative blots are shown. (E) Quantification of the Western blotting data shown in D. Data are mean ± SEM of 4 independent experiments. *P < 0.05; NS, no statistically significant difference (B, Kruskal-Wallis test; C, 2-tailed Student’s t test; E, 2-way ANOVA followed by Tukey’s post hoc test). See complete unedited blots in the supplemental material.
Barr1 is required for SU/Epac2-mediated activation of Rap1. Since activated Epac2 functions as a Rap1 GEF, we next examined whether Barr1 knockdown in MIN6-K8 cells affected the ability of glibenclamide (0.1 μM) to activate endogenous Rap1. The efficient knockdown of both Barr1 mRNA and Barr1 protein was confirmed by real-time reverse transcription PCR (qRT-PCR) (Figure 4C) and immunoblotting studies (Supplemental Figure 15), respectively. To detect activated Rap1 (Rap1-GTP), we used a GST-RalGDS-RBD fusion protein that specifically binds Rap1-GTP, followed by the detection of Rap1-GTP by Western blotting. We found that glibenclamide-dependent Rap1 activation was greatly reduced after Barr1 knockdown (Figure 4, D and E), supporting the concept that efficient SU activation of Epac2/Rap1 signaling requires the presence of Barr1. Our data are consistent with a model in which Barr1 forms a complex with Epac2 that is stabilized by SUs such as glibenclamide. The formation of this complex then promotes enhanced Rap1 activity and insulin secretion. Our observation that Barr1 is required for SU/Epac2-mediated activation of Rap1 provides an explanation for the previous finding that SUs were unable to activate purified Epac2 directly (27).
Conclusion. We demonstrated that Barr1 is required for efficient SU-stimulated insulin secretion from pancreatic β cells. Our data indicate that Barr1 promotes SU-induced insulin secretion by binding to Epac2, thus enhancing Epac2-induced Rap1 activation. These results suggest that strategies aimed at promoting β cell Barr1 signaling may prove useful for the development of efficacious antidiabetic drugs.
-
Methods
Detailed methods are described in Supplemental Methods. See complete unedited blots in the supplemental material.
Statistics. Data are mean ± SEM. Statistical differences were determined using either Student’s t test (2-tailed) or 2-way ANOVA followed by Tukey’s post hoc test, as appropriate. P values less than 0.05 were considered significant.
Study approval. All animal studies were approved by the National Institute of Diabetes and Digestive and Kidney Diseases Institutional Animal Care and Use Committee.
-
Author contributions
LFB, MR, and JW designed and conceived the experiments. LFB, MR, LZ, and YC performed and analyzed experiments. The lab of WC generated the floxed Barr1 mice and provided critical know-how regarding the use of these mice. XC, FCM, and VVG provided novel reagents and helpful advice throughout the course of this study. LFB and JW wrote the manuscript.
-
Acknowledgments
This research was funded by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases (to LFB, MR, LZ, YC, and JW), NIH grant R35 GM122491 (to VVG), and NIGMS grant R35 GM122536 (to XC). LFB received a 2-year stipend through a joint program between NIH and the Brazilian National Council for Scientific and Technological Development (CNPq). The adenovirus coding for FLAG- Epac2 was a gift by George Holz (SUNY Upstate Medical University, Syracuse, NY). Robert J. Lefkowitz and his colleagues at Duke University provided the floxed Barr1 mice and the whole-body Barr1 and Barr2 KO mice. The purification of bovine Barr1 was carried out by Sergey A. Vishnivetskiy (Vanderbilt University, Nashville, TN). We thank Oksana Gavrilova (NIDDK Mouse Metabolic Core) for advice and experimental support. Joana Almaça and Alejandro Caicedo (University of Miami Miller School of Medicine, Miami, FL) provided LFB with training and advice regarding the islet perifusion studies.
Address correspondence to: Jürgen Wess, Laboratory of Bioorganic Chemistry, Molecular Signaling Section, National Institute of Diabetes and Digestive and Kidney Diseases, Bldg. 8A, Room B1A-05, 8 Center Drive, Bethesda, Maryland 20892, USA. Phone: 301.402.3589; E-mail: jurgenw@niddk.nih.gov.
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(9):3732–3737.https://doi.org/10.1172/JCI126309.
-
References
- Zhao J, Pei G. Arrestins in metabolic regulation. Prog Mol Biol Transl Sci. 2013;118:413–427.
- Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3(9):639–650.
- Gurevich VV, Gurevich EV. Overview of different mechanisms of arrestin-mediated signaling. Curr Protoc Pharmacol. 2014;67:Unit 2.10.1–Unit 2.10.9.View this article via: PubMed Google Scholar
- Shukla AK, Xiao K, Lefkowitz RJ. Emerging paradigms of β-arrestin-dependent seven transmembrane receptor signaling. Trends Biochem Sci. 2011;36(9):457–469.
- Zhu L, et al. β-arrestin-2 is an essential regulator of pancreatic β-cell function under physiological and pathophysiological conditions. Nat Commun. 2017;8:14295. View this article via: PubMed Google Scholar
- Sola D, et al. Sulfonylureas and their use in clinical practice. Arch Med Sci. 2015;11(4):840–848.View this article via: PubMed Google Scholar
- Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129(10):2447–2457.View this article via: PubMed Google Scholar
- Fu A, et al. Loss of Lkb1 in adult beta cells increases beta cell mass and enhances glucose tolerance in mice. Cell Metab. 2009;10(4):285–295.
- Kim J, et al. β-arrestin 1 regulates β2-adrenergic receptor-mediated skeletal muscle hypertrophy and contractility. Skelet Muscle. 2018;8(1):39.
- Gautam D, et al. A critical role for beta cell M3 muscarinic acetylcholine receptors in regulating insulin release and blood glucose homeostasis in vivo. Cell Metab. 2006;3(6):449–461.
- Nakajima K, Jain S, Ruiz de Azua I, McMillin SM, Rossi M, Wess J. Minireview: Novel aspects of M3 muscarinic receptor signaling in pancreatic β-cells. Mol Endocrinol. 2013;27(8):1208–1216.
- Ahrén B. Islet G protein-coupled receptors as potential targets for treatment of type 2 diabetes. Nat Rev Drug Discov. 2009;8(5):369–385.
- Drucker DJ. Mechanisms of action and therapeutic application of glucagon-like peptide-1. Cell Metab. 2018;27(4):740–756.
- Ruiz de Azua I, et al. RGS4 is a negative regulator of insulin release from pancreatic beta-cells in vitro and in vivo. Proc Natl Acad Sci U S A. 2010;107(17):7999–8004.
- Lamont BJ, Li Y, Kwan E, Brown TJ, Gaisano H, Drucker DJ. Pancreatic GLP-1 receptor activation is sufficient for incretin control of glucose metabolism in mice. J Clin Invest. 2012;122(1):388–402.
- Smith EP, et al. The role of β cell glucagon-like peptide-1 signaling in glucose regulation and response to diabetes drugs. Cell Metab. 2014;19(6):1050–1057.
- Sonoda N, Imamura T, Yoshizaki T, Babendure JL, Lu JC, Olefsky JM. Beta-arrestin-1 mediates glucagon-like peptide-1 signaling to insulin secretion in cultured pancreatic beta cells. Proc Natl Acad Sci U S A. 2008;105(18):6614–6619.
- Proks P, Reimann F, Green N, Gribble F, Ashcroft F. Sulfonylurea stimulation of insulin secretion. Diabetes. 2002;51(Suppl 3):S368–S376.View this article via: PubMed Google Scholar
- Zhang CL, et al. The cAMP sensor Epac2 is a direct target of antidiabetic sulfonylurea drugs. Science. 2009;325(5940):607–610.
- Hinke SA. Epac2: a molecular target for sulfonylurea-induced insulin release. Sci Signal. 2009;2(85):pe54. View this article via: PubMed Google Scholar
- Takahashi T, et al. Antidiabetic sulfonylureas and cAMP cooperatively activate Epac2A. Sci Signal. 2013;6(298):ra94. View this article via: PubMed Google Scholar
- Herbst KJ, Coltharp C, Amzel LM, Zhang J. Direct activation of Epac by sulfonylurea is isoform selective. Chem Biol. 2011;18(2):243–251.
- Shibasaki T, et al. Essential role of Epac2/Rap1 signaling in regulation of insulin granule dynamics by cAMP. Proc Natl Acad Sci U S A. 2007;104(49):19333–19338.
- Mangmool S, Shukla AK, Rockman HA. Beta-arrestin-dependent activation of Ca(2+)/calmodulin kinase II after beta(1)-adrenergic receptor stimulation. J Cell Biol. 2010;189(3):573–587.
- Tsalkova T, et al. Isoform-specific antagonists of exchange proteins directly activated by cAMP. Proc Natl Acad Sci U S A. 2012;109(45):18613–18618.
- Iwasaki M, Minami K, Shibasaki T, Miki T, Miyazaki J, Seino S. Establishment of new clonal pancreatic β-cell lines (MIN6-K) useful for study of incretin/cyclic adenosine monophosphate signaling. J Diabetes Investig. 2010;1(4):137–142.View this article via: PubMed Google Scholar
- Tsalkova T, Gribenko AV, Cheng X. Exchange protein directly activated by cyclic AMP isoform 2 is not a direct target of sulfonylurea drugs. Assay Drug Dev Technol. 2011;9(1):88–91.
-
Version history
- Version 1 (June 11, 2019): In-Press Preview
- Version 2 (August 5, 2019): Electronic publication
- Version 3 (September 3, 2019): Print issue publication



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)