Advertisement
Research ArticleEndocrinology Free access | 10.1172/JCI121679
Fbxo22-mediated KDM4B degradation determines selective estrogen receptor modulator activity in breast cancer
Yoshikazu Johmura,1 Ichiro Maeda,2 Narumi Suzuki,1 Wenwen Wu,3 Atsushi Goda,2 Mariko Morita,4 Kiyoshi Yamaguchi,4 Mizuki Yamamoto,5 Satoi Nagasawa,6 Yasuyuki Kojima,6 Koichiro Tsugawa,6 Natsuko Inoue,7 Yasuo Miyoshi,7 Tomo Osako,8 Futoshi Akiyama,8 Reo Maruyama,9 Jun-ichiro Inoue,5 Yoichi Furukawa,4 Tomohiko Ohta,3 and Makoto Nakanishi1
1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by Johmura, Y. in: PubMed | Google Scholar
1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by
Maeda, I.
in:
PubMed
|
Google Scholar
|

1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by Suzuki, N. in: PubMed | Google Scholar
1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by Wu, W. in: PubMed | Google Scholar
1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by Goda, A. in: PubMed | Google Scholar
1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by Morita, M. in: PubMed | Google Scholar
1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by
Yamaguchi, K.
in:
PubMed
|
Google Scholar
|

1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by Yamamoto, M. in: PubMed | Google Scholar
1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by Nagasawa, S. in: PubMed | Google Scholar
1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by Kojima, Y. in: PubMed | Google Scholar
1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by Tsugawa, K. in: PubMed | Google Scholar
1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by Inoue, N. in: PubMed | Google Scholar
1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by
Miyoshi, Y.
in:
PubMed
|
Google Scholar
|

1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by
Osako, T.
in:
PubMed
|
Google Scholar
|

1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by Akiyama, F. in: PubMed | Google Scholar
1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by Maruyama, R. in: PubMed | Google Scholar
1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by Inoue, J. in: PubMed | Google Scholar
1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by Furukawa, Y. in: PubMed | Google Scholar
1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by Ohta, T. in: PubMed | Google Scholar
1Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
2Department of Pathology St. Marianna University School of Medicine, Kawasaki, Japan.
3Department of Translational Oncology, St. Marianna University Graduate School of Medicine, Kawasaki, Japan.
4Division of Clinical Genome Research, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
5Division of Cellular and Molecular Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
6Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Japan.
7Division of Breast and Endocrine Surgery, Department of Surgery, Hyogo College of Medicine, Hyogo, Japan.
8Department of Pathology, The Cancer Institute Hospital, and
9Project for Cancer Epigenomics, the Cancer Institute of the Japanese Foundation for Cancer Research, Tokyo, Japan.
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
Find articles by
Nakanishi, M.
in:
PubMed
|
Google Scholar
|

Published November 12, 2018 - More info
J Clin Invest. 2018;128(12):5603–5619. https://doi.org/10.1172/JCI121679.
Copyright © 2018, American Society for Clinical Investigation
Received: April 17, 2018; Accepted: September 27, 2018
-
Abstract
The agonistic/antagonistic biocharacter of selective estrogen receptor modulators (SERMs) can have therapeutic advantages, particularly in the case of premenopausal breast cancers. Although the contradictory effects of these modulators have been studied in terms of crosstalk between the estrogen receptor α (ER) and coactivator dynamics and growth factor signaling, the molecular basis of these mechanisms is still obscure. We identify a series of regulatory mechanisms controlling cofactor dynamics on ER and SERM function, whose activities require F-box protein 22 (Fbxo22). Skp1, Cullin1, F-box–containing complex (SCFFbxo22) ubiquitylated lysine demethylase 4B (KDM4B) complexed with tamoxifen-bound (TAM-bound) ER, whose degradation released steroid receptor coactivator (SRC) from ER. Depletion of Fbxo22 resulted in ER-dependent transcriptional activation via transactivation function 1 (AF1) function, even in the presence of SERMs. In living cells, TAM released SRC and KDM4B from ER in a Fbxo22-dependent manner. SRC release by TAM required Fbxo22 on almost all ER-SRC–bound enhancers and promoters. TAM failed to prevent the growth of Fbxo22-depleted, ER-positive breast cancers both in vitro and in vivo. Clinically, a low level of Fbxo22 in tumor tissues predicted a poorer outcome in ER-positive/human epidermal growth factor receptor type 2–negative (HER2-negative) breast cancers with high hazard ratios, independently of other markers such as Ki-67 and node status. We propose that the level of Fbxo22 in tumor tissues defines a new subclass of ER-positive breast cancers for which SCFFbxo22-mediated KDM4B degradation in patients can be a therapeutic target for the next generation of SERMs.
-
Introduction
Estrogen receptor α (ER) belongs to the nuclear receptor superfamily, and ligand-bound ER modulates the rate of transcriptional initiation of various downstream target genes through synergistic action between transactivation function 1 (AF1) and transactivation function 2 (AF2) (1, 2). This modulation is mediated at least in part through recruitment of a variety of coactivators or corepressors, thus leading to alterations in the state of chromatin organization at target promoters (3–5). ER and its coactivators associate with estrogen-responsive elements during estrogen treatment. In contrast, antagonist-bound ER forms a complex with corepressors, instead of coactivators, at promoters (4).
Breast cancer is the most frequently diagnosed cancer in women. Approximately 70% of breast cancers are positive for ER, and TAM is the standard drug for treatment of ER-positive breast cancer, especially for premenopausal women (6, 7). Treatment with TAM as an adjuvant decreases the annual breast cancer mortality rate by approximately 30% (8, 9). However, up to 25% of patients with early-stage breast cancer treated with TAM experience a relapse of the disease within 15 years (10). Hence, modification of the treatment is absolutely required for some populations. TAM is classified as a type of selective ER modulator (SERM). In some tissues, SERMs act like estrogens, but in other tissues, estrogen action is blocked by SERMs as opposed to selective ER degraders (SERDs), which act as full antagonists including fulvestrant (11, 12). Selective modulator functions of SERMs are therefore beneficial for treating premenopausal ER-positive breast cancers and also reduce such harmful effects as an increased susceptibility to osteoporosis and coronary heart disease (13). This selective modulation has been reported to stem from a complete blockade of AF2 function, but in a context specific to the function of AF1 (14–16). Previous studies have demonstrated that the relative expression of coactivators and corepressors in cells contributes to the selective modulation of ER function (17–20). In patients with breast cancer, a high level of steroid receptor coactivator 3 (SRC-3) expression correlates with TAM resistance (21). Overexpression of SRC-3 and human epidermal growth factor receptor type 2 (HER2) in MCF7 cells converts TAM from an ER antagonist into an ER agonist (22). However, since overexpression of SRCs, per se, failed to suppress the antagonistic action of SERMs on estrogen response elements (EREs) (17), and since some ER-positive and HER2-negative breast cancers also show a resistance to TAM, it is likely that there are unidentified mechanism(s) underlying cofactor dynamics on ER that determine SERM function.
The histone lysine demethylase 4 (KDM4) family plays an essential role in the regulation of transcription by changing chromatin architecture in response to steroid hormones (23). For example, KDM4C is an essential coactivator of androgen receptor–mediated transcription and androgen-dependent cellular growth in prostate cancers (24, 25). More recently, KDM4B has been reported to be required for ER-mediated transcription and proliferation of breast cancer cells through direct interaction with ER and through KDM4B lysine demethylase activity (26–29). While KDM4B is a bona fide target of ER, it also transcriptionally upregulates ER expression (26), creating a positive feedback loop between KDM4B and ER.
Fbxo22 is not yet well characterized, but it is known to be an F-box protein that contains 3 functional domains: F-box, F-box and intracellular signal transduction proteins FIST-N, and FIST-C. It was originally reported to be a transcriptional target of p53 (30), and later to form a complex with KDM4A, whose degradation regulates histone H3 methylation at lysines 9 and 36 (31). We have recently identified the Skp1, Cullin1, F-box–containing Fbxo22–KDM4A (SCFFbxo22–KDM4A) complex as an essential component in the maintenance of the senescent state, in which it functions as an E3 ubiquitin ligase for methylated p53 (32). However, to date, only a subset of SCFFbxo22 targets has emerged.
In this study, we unveiled the essential role of Fbxo22 in the antagonistic action of SERMs. We found that this action is mediated by the selective degradation of KDM4B complexed with TAM-bound ER. Loss of Fbxo22 permits ER-dependent transcription and recruitment of coactivators on almost all ER-responsive promoters, even in the presence of SERMs. Our T47D xenograft model also supports the role of Fbxo22 in the antagonistic activity in vivo. Finally, we found that a low level of Fbxo22 predicts a poorer outcome in patients with ER-positive breast cancer, confirming the clinical and physiological importance of Fbxo22 in the pharmacological actions of SERMs.
-
Results
Ubiquitylation and degradation of KDM4B is essential for cofactor dynamics on ER and determines the antagonistic activity of TAM. Given that E2-induced transcriptional activity in MCF7 cells is regulated by proteasome-mediated degradation (33, 34), we speculated that the antagonistic activity of TAM might also be regulated by proteasome-mediated protein degradation. First, we determined the kinetics of the antagonistic activity of 4-hydroxytamoxifen (4-OHT). E2-depleted MCF7 cells were stimulated by the addition of E2. Estrogen signaling was then antagonized by treatment with 4-OHT six hours after E2 stimulation. We found that the levels of transcripts of GREB1 and EBAG9, known ER targets, increased and reached a maximum 4 hours after the addition of E2. The levels then decreased and reached a minimum 6 hours after the addition of 4-OHT (Figure 1A), indicating that estrogen signaling is rapidly antagonized after TAM treatment. We then examined whether the proteasome inhibitor MG132 affected the antagonistic activity of 4-OHT on ER-mediated transcription in MCF7 cells. After 4-OHT treatment, we observed that the EBAG9 and GREB1 transcripts were maintained at higher levels in the presence of MG132 than those in the absence of MG132 (Figure 1A). ER formed a complex with KDM4B and SRC-3, a known coactivator of steroid receptors, as well as with p300 (35, 36), after E2 treatment (Figure 1B). This complex dissociated after 4-OHT addition, and, in turn, ER formed a complex with the N-CoR repressor and HDAC3. Treatment with MG132 suppressed these cofactor dynamics. Similar results were obtained when we treated T47D cells with 4-OHT (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI121679DS1). Removal of E2 from E2-containing medium abolished the E2-induced transcriptional activation of both genes (Figure 1C). Again, treatment with MG132 compromised this abrogation and suppressed the dissociation of KDM4B, SRC-3, and p300 from ER by the removal of E2 (Figure 1D). Given these results, we speculated that selective degradation of KDM4B complexed with ER by proteasomes might trigger ER cofactor dynamics. As expected, we found that KDM4B depletion resulted in the dissociation of SRC-3 and p300 from ER, even in the presence of E2 (Figure 1E). Together, our results suggested that selective degradation of KDM4B on ER by proteasomes is likely to trigger cofactor dynamics and facilitate the antagonistic activity of TAM in ER-positive breast cancer cells.
 Figure 1
Figure 1Proteasome-dependent protein degradation is required for the antagonistic activity of TAM. (A) Experimental outline (top). MCF7 cells starved of E2 for 72 hours were cultured in medium containing E2 (10 nM) for 6 hours and then incubated in medium containing 4-OHT (100 nM) with or without MG132 (10 μg/ml). Total RNA from the treated cells collected at the indicated time points was subjected to qRT-PCR analysis using the indicated primers. Data are presented as the mean ± SD of 3 independent experiments. ****P < 0.001 and ***P < 0.005, by 2-tailed Student’s t test. (B) Nuclear extracts (NEs) of cells treated as in A were collected at 12 hours and immunoprecipitated using the indicated antibodies and then subjected to immunoblotting. (C) MCF7 cells starved of E2 for 72 hours were cultured with medium containing E2 (10 nM) for 18 hours (E2), or for 6 hours with or without MG132 (10 μg/ml) and then starved of E2 (E2-dep) for 12 hours. Total RNA from the treated cells was subjected to qRT-PCR analysis using the indicated primers. Data are presented as the mean ± SD of 3 independent experiments. *P < 0.05, by 2-tailed Student’s t test. (D) Nuclear extracts of cells treated as described in C were immunoprecipitated using the indicated antibodies and subjected to immunoblotting. (E) MCF7 cells expressing the indicated doxycycline-inducible shRNAs (Dox-shRNA-MCF7 cells) were starved of E2 in the presence of doxycycline (1 μg/ml) for 72 hours and then treated with E2 (10 nM) for 6 hours. Nuclear extracts were immunoprecipitated using the indicated antibodies and subjected to immunoblotting.
Fbxo22 forms a ternary complex with KDM4B and ER, depending on the type of ligand. We next examined a factor that selectively degrades KDM4B complexed with ER. Given that Fbxo22 has been reported to be related to KDM4A function (31), we examined whether it regulates KDM4B as well. Although the steady-state levels of KDM4A, 4C, and 4D were comparable between WT and Fbxo22-depleted cells, we found that the levels of KDM4B markedly increased (Figure 2A), whereas the levels of KDM4B mRNA did not vary (Supplemental Figure 2A). KDM4B protein was more stable in Fbxo22-depleted cells than in the control MCF7 and T47D cells (Figure 2B and Supplemental Figure 2B). To address the question of whether SCFFbxo22 ubiquitylates KDM4B complexed with ER, we first examined complex formation between ER and Fbxo22. When FLAG-HA–tagged Fbxo22 (FH-Fbxo22) was expressed in MCF7 cells in the presence of MG132, a sequential immunopurification using anti-FLAG and anti-HA antibodies revealed that ER interacted with FH-Fbxo22 (Figure 2C). The endogenous interaction between ER, Fbxo22, and KDM4B was also confirmed in the presence of MG132 in MCF7 and T47D cells (Figure 2D and Supplemental Figure 2C). Importantly, we observed that the interaction between ER and KDM4B was markedly enhanced when endogenous Fbxo22 was depleted in the absence of MG132 in MCF7 and T47D cells (Figure 2, E and F, and Supplemental Figure 2, D and E ). Given that Fbxo22 possesses 3 distinct functional domains, F-box, FIST-N, and FIST-C (32), we speculated that Fbxo22 forms a multimeric complex with ER and KDM4B. Using Fbxo22 mutants lacking the FIST-N or FIST-C domains, we found that ER and KDM4B bound to FIST-N and FIST-C domains in MCF7 and T47D cells, respectively (Figure 2G and Supplemental Figure 2F). In addition, when FLAG-tagged Fbxo22 was expressed in MCF7 and T47D cells, sequential immunoprecipitation confirmed a ternary complex comprising FLAG-Fbxo22, ER, and KDM4B in the presence of MG132 (Figure 2H and Supplemental Figure 2G).
 Figure 2
Figure 2Fbxo22 forms a ternary complex with ER and KDM4B in a ligand type–dependent manner in MCF7 cells. (A) MCF7 cells expressing the indicated doxycycline-inducible shRNAs (Dox-shRNA-MCF7 cells) were treated with doxycycline (1 μg/ml). At the indicated time points, the lysates were subjected to immunoblotting. (B) The indicated Dox-shRNA-MCF7 cells, in the presence of doxycycline (1 μg/ml) for 24 hours, were treated with 50 μg/ml cycloheximide (CHX) and analyzed as in A. The relative KDM4B intensities were determined using ImageJ software. Data are presented as the mean ± SD of 3 independent experiments. ****P < 0.001, by 2-tailed Student’s t test. (C) Dox-FLAG-HA-Fbxo22-MCF7 cells, in the presence or absence of doxycycline (1 μg/ml) for 48 hours, were treated with MG132 (10 μg/ml) for 4 hours. The whole-cell extracts (WCEs) were sequentially immunoprecipitated using anti-FLAG M2 gel and anti-HA gel and then subjected to immunoblotting. (D) MCF7 cells were treated with MG132 (10 μg/ml) for 4 hours. WCEs were immunoprecipitated and subjected to immunoblotting using the indicated antibodies. (E and F) The indicated Dox-shRNA-MCF7 cells were incubated with doxycycline (1 μg/ml) for 48 hours and analyzed as in D. (G) MCF7 cells, Dox-WT FLAG-Fbxo22 (WT) cells, or mutant cells lacking FIST-N (ΔFN) or FIST-C (ΔFC) were treated as in C. WCEs were immunoprecipitated using anti-FLAG M2 affinity gel and subjected to immunoblotting. (H) Dox-FLAG-Fbxo22-MCF7 cells were treated as in C. WCEs were sequentially immunoprecipitated with anti-FLAG M2 gel and anti-ER antibodies and subjected to immunoblotting. Dox-FLAG-Fbxo22-MCF7 cells were starved in E2-depleted medium with or without doxycycline (1 μg/ml) for 72 hours and treated with or without 0.1 nM, 1 nM, or 10 nM E2 (I) or E2 (10 nM) and/or 1 nM, 10 nM, or 100 nM 4-OHT (J) in the presence of MG132 (10 μg/ml) for 6 hours. WCEs were immunoprecipitated using the indicated antibodies and subjected to immunoblotting.
We then examined the effects of an agonist or antagonist binding to ER in Fbxo22 interaction. FLAG-Fbxo22 binding to ER was decreased by the addition of E2 in a dose-dependent manner in the presence of MG132, although its binding to KDM4B was not affected in MCF7 and T47D cells (Figure 2I and Supplemental Figure 2H). We observed that a reduction of FLAG-Fbxo22 binding to ER in the presence of E2 and MG132 was reversed by the addition of 4-OHT in a dose-dependent manner (Figure 2J and Supplemental Figure 2I). These results suggest that Fbxo22 preferentially binds to unliganded or 4-OHT–bound ER, but not to E2-bound ER.
SCFFbxo22 ubiquitylates KDM4B complexed with ER in a manner dependent on the type of ligand. Although the level of KDM4B was not affected by ectopic expression of either Fbxo22 or ER, the expression of both proteins decreased KDM4B expression levels (Figure 3A), thus suggesting that SCFFbxo22 might specifically ubiquitylate KDM4B complexed with ER for subsequent degradation. To examine this possibility, we first examined whether the F-box domain of Fbxo22 or the catalytic activity of KDM4B plays a role in the regulation of KDM4B stability. Although reintroduction of WT Fbxo22 reversed KDM4B stabilization in Fbxo22-KO cells, that of a mutant Fbxo22 lacking the F-box (ΔFbox) failed to do so (Supplemental Figure 3A). In addition, Fbxo22 destabilized both WT KDM4B and a catalytic mutant (H189A) of KDM4B to a similar extent (Supplemental Figure 3B). Taken together, the results suggest that the F-box (ubiquitylation activity) of Fbxo22, but not the catalytic activity of KDM4B, is essential for SCFFbxo22-dependent regulation of KDM4B stability. We then performed an in vivo ubiquitylation assay under denaturing conditions. Although Fbxo22-induced ubiquitylation of KDM4B was weakly detected without ectopic expression of ER, this signal was enhanced by coexpression of ER in a dose-dependent manner (Figure 3B). Together, these results suggested that SCFFbxo22 ubiquitylates KDM4B complexed with ER for proteasomal degradation. Given that Fbxo22 preferentially binds to unliganded or 4-OHT–bound ER, ubiquitylation of KDM4B complexed with ER by SCFFbxo22 probably depends on the type of ligand bound to ER. We observed that the ubiquitylation of KDM4B complexed with ER was compromised by the addition of E2 and that this reduced ubiquitylation was recovered by adding 4-OHT (Figure 3C). Taken together, SCFFbxo22 preferentially ubiquitylates KDM4B complexed with unliganded or 4-OHT–bound ER.
 Figure 3
Figure 3SCFFbxo22 preferentially ubiquitylates KDM4B complexed with unliganded or 4-OHT–bound ER. (A) Lysates from MCF7 cells expressing Fbxo22 and/or ER were subjected to immunoblotting using the indicated antibodies. Numbers shown at the bottom of the KDM4B blot indicate relative signal intensities. (B) Fbxo22-KO HeLa cells were transfected with the indicated plasmids, treated with MG132, and lysed under denaturing conditions. WCEs were subjected to StrepTactin (Strep) pulldown, followed by immunoblotting. (C) Fbxo22-KO HeLa cells expressing the same genes as indicated in B were treated in the presence or absence of E2 and 4-OHT. WCEs were subjected to StrepTactin pulldown, followed by immunoblotting. St2-KDM4B, tandem strep-II–tagged KDM4B.
SCFFbxo22-mediated degradation of KDM4B is required for the antagonistic activity of SERMs. On the basis of the observation that the antagonistic activity of 4-OHT against E2 signaling requires proteasome-mediated protein degradation (Figure 1, A–E), SCFFbxo22-mediated ubiquitylation of KDM4B complexed with ER for degradation is probably essential for this antagonistic activity through cofactor dynamics. Treatment with 4-OHT in E2-stimulated MCF7 and T47D cells resulted in strong suppression of EBAG9 and GREB1 transcription, whereas the same antagonistic activity of 4-OHT was suppressed in E2-stimulated MCF7 and T47D cells lacking Fbxo22 (Figure 4A and Supplemental Figure 4A). Likewise, transcriptional suppression of ER by E2 removal also required Fbxo22 (Supplemental Figure 5A). Reintroduction of WT Fbxo22 reversed the antagonistic activity of 4-OHT (Supplemental Figure 6A). Depletion of Fbxo22 and its reintroduction, per se, did not affect transcriptional induction of ER target genes by E2. After E2 stimulation, ER formed a complex with KDM4B and SRC-3 in both control and Fbxo22-depleted cells. After treatment with 4-OHT, KDM4B and SRC-3 dissociated from ER, whereas N-CoR associated with ER in control MCF7 and T47D cells (Figure 4B and Supplemental Figure 4B). In contrast, we did not observe these cofactor dynamics in Fbxo22-depleted cells (Figure 4B and Supplemental Figure 4B), and we observed similar results after E2 depletion (Supplemental Figure 5B). KDM4B depletion also resulted in a marked suppression of E2-induced transcriptional induction of EBAG9 and other ER target genes, independent of the presence of Fbxo22, confirming the epistasis of KDM4B in regulating the antagonistic activity of 4-OHT (Supplemental Figure 6B).
 Figure 4
Figure 4Antagonistic activity of 4-OHT requires Fbxo22 via AF1 activity in MCF7 cells. (A) Experimental outline (top). Dox–shFbxo22–MCF7 cells were starved of E2 in the presence or absence of doxycycline (1 μg/ml) for 72 hours, treated with E2 (10 nM) for 6 hours, and then treated with 4-OHT (100 nM). Total RNA from the treated cells at the indicated time points was subjected to qRT-PCR analysis. Data are presented as the mean ± SD of 3 independent experiments. *P < 0.05 and ****P < 0.001, by 2-tailed Student’s t test. (B) Nuclear extracts from the cells treated as in A at 12 hours were immunoprecipitated using the indicated antibodies and subjected to immunoblotting. (C) U2OS cells expressing WT ER or its Δ44 mutant were treated and analyzed as in A. ****P < 0.001, by 2-tailed Student’s t test.
As mentioned above, the transcriptional activity of ER is elicited by AF1 and AF2. Thus, we determined whether ER activity in Fbxo22-depleted cells in the presence of 4-OHT depends on AF1 and/or AF2. U2OS cells, with or without Fbxo22, that expressed full-length WT ER or its Δ44 mutant lacking AF1 activity were stimulated with E2 and then treated with 4-OHT (Supplemental Figure 7A). We found that the antagonistic activity of 4-OHT was suppressed in E2-stimulated U2OS cells expressing WT ER in the absence of Fbxo22, while this activity was maintained in those expressing Δ44 ER, also in the absence of Fbxo22 (Figure 4C and Supplemental Figure 7B). Thus, the results suggest that ER activity in the presence of 4-OHT in cells lacking Fbxo22 depends on AF1 activity. The role of Fbxo22 in antagonistic activity against ER signaling appeared to be specific to SERMs, but not SERDs, showing that Fbxo22 depletion had a similar effect or no effect at all on the antagonistic activity of toremifene and fulvestrant, respectively (Supplemental Figure 8A). The cofactor dynamics on treating cells with toremifene required Fbxo22, but the dynamics with fulvestrant did not (Supplemental Figure 8B). Thus, these results suggested that SCFFbxo22-mediated degradation of KDM4B on the ER is required for cofactor dynamics specifically upon SERM treatment.
In order to further confirm the essential role of Fbxo22 in ligand-mediated assembly of coactivators and ER, we examined the real-time cellular dynamics of ER-coactivator complexes in living cells. We used a CFP-tagged lac repressor–ER chimera (CFP-LacER) in live cells to discretely immobilize ER on stably integrated Lac operator arrays as a means to examine the recruitment of YFP–SRC-1 as well as KDM4B. CFP-LacER was reported to retain the ability to activate transcription in an agonist-dependent manner (37). Agonist- or antagonist-dependent ER–SRC-1 interactions were analyzed using U2OS-LacO-I-SceI TetO cells (38) expressing CFP-LacER and YFP–SRC-1. In the absence of E2, we found that most of the CFP-LacER localized at LacO foci in either control or Fbxo22-depleted cells, whereas YFC–SRC-1 did not (Supplemental Figure 9, A and B). In the presence of E2, more than 70% of CFP-LacER foci colocalized with those of YFP–SRC-1 in control cells, but failed to do so in cells in the additional presence of 4-OHT. In contrast, more than 70% of CFP-LacER foci colocalized with those of YFP–SRC-1 in the presence of E2 in Fbxo22-depleted cells, and nearly 50% of CFP-LacER foci still retained colocalization with YFP–SRC-1 in the presence of both E2 and 4-OHT.
We then examined the colocalization of FLAG-KDM4B with CFP-LacER and YFP–SRC-1 foci using U2OS-LacO-I-SceI TetO cells expressing CFP-LacER, YFP–SRC-1, and FLAG-KDM4B. An immunofluorescence analysis using anti-FLAG antibodies demonstrated that FLAG-KDM4B also colocalized with CFP-LacER and YFP–SRC-1 foci in control and Fbxo22-depleted cells in the presence of E2, whereas it failed to do so in control cells in the presence of both E2 and 4-OHT (Figure 5, A and B). Again, FLAG-KDM4B colocalized with CFP-LacER and YFP–SRC-1 foci in Fbxo22-depleted cells in the presence of both E2 and 4-OHT. Taken together, these results also indicate the essential role of Fbxo22 in cofactor dynamics upon shutdown of E2 signaling by 4-OHT.
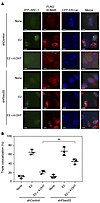 Figure 5
Figure 5Essential role of Fbxo22 in 4-OHT–induced dissociation of SRC-1 from ER in living cells. (A) The indicated Dox-shRNAs-U2OS-LacO-I-SceI-TetO cells expressing FLAG-KDM4B were transfected with YFP–SRC-1 and CFP-ERα-Lac plasmids, followed by treatment with doxycycline (1 μg/ml) in E2-depleted medium for 72 hours. These cells were then treated with or without E2 (10 nM) and/or 4-OHT (100 nM) for 2 hours and fixed with 4% formaldehyde. The resultant cells were subjected to immunostaining with anti-FLAG antibody. Representative images are shown. Scale bars: 10 μm. (B) Cells positive for triple colocalization of CFP-ERα-Lac, YFP–SRC-1, and FLAG-KDM4B foci were counted. Data are presented as the mean ± SD of 3 independent experiments. **P < 0.01, by 2-tailed Student’s t test.
Fbxo22 is required for 4-OHT–mediated SRC-3 release from almost all ER-SRC-3–bound genomic regions. We then mapped genome-wide ER- and SRC-3–binding events by ChIP-sequencing (ChIP-Seq) analysis. We performed the analysis using MCF7 and Fbxo22-depleted MCF7 cells. Cells were estrogen starved for 72 hours and subsequently stimulated for 2 hours with E2 (10 nM) or E2 plus TAM. As a result, we identified a total of 12,645 and 23,216 enriched ER peaks in MCF7 and Fbxo22-depleted MCF7 cells, respectively, in cells treated with E2, and 26,751 and 19,924 enriched ER peaks in MCF7 and Fbxo22-depleted MCF7 cells, respectively, in cells treated with E2 plus 4-OHT. These results suggested that 4-OHT is unlikely to affect the interaction between ER and its target regions. The above 4 data sets shared 8,528 overlapping ER peaks. We then identified 1,723 and 5,572 enriched SRC-3 peaks in MCF7 and Fbxo22-depleted MCF7 cells, respectively, in the presence of E2. These 2 data sets shared 469 SRC-3 peaks, and nearly 90% of these peaks (n = 410) overlapped the ER peaks (Figure 6A). To determine the amount of SRC-3 that was associated with ER, we focused on the 410 overlapping SRC-3 peaks. The box-and-whisker plot clearly showed a significant decrease in SRC-3 associated with ER in MCF7 cells treated with E2 plus 4-OHT, whereas this decrease was not observed in Fbxo22-depleted MCF7 cells (Figure 6B). The heatmap showed a marked decrease in SRC-3 sequencing reads across the 410 regions in MCF7 cells treated with E2 plus 4-OHT compared with those treated with E2 alone. In contrast, the densities of SRC-3 sequencing reads were not affected by the addition of 4-OHT to Fbxo22-depleted MCF7 cells (Figure 6C). Indeed, treatment of MCF7 and Fbxo22-depleted MCF7 cells with E2 promoted ER recruitment to the promoter regions of the GREB1 and IGFBP4 genes (Figure 6D). The ER was retained on these promoters after treatment with E2 plus 4-OHT in both cells. SRC-3 was also recruited to the same regions as those of ER bound on the promoters in the presence of E2 in both MCF7 and Fbxo22-depleted MCF7 cells. In contrast, in MCF7 cells, SRC-3 dissociated from the promoters when the cells were treated with E2 plus 4-OHT, whereas in Fbxo22-depleted MCF7 cells, it was retained on the promoters under the same conditions. Taken together, these results clearly indicate that Fbxo22 plays a critical and universal role in TAM-induced dissociation of SRC-3 from almost all ER- and SRC-3–binding genomic regions.
 Figure 6
Figure 6Genome-wide analysis revealed that TAM-mediated dissociation of SRC-3 from ER requires Fbxo22 on almost all ER- and SRC-3–binding sites. (A) ChIP binding site counts and proximity relationships. Venn diagram of binding sites for ER with sequence centers within 0.1 kb of each other were identified as a shared peak within 4 data sets (MCF7 cells with E2 and E2 plus 4-OHT, and Fbxo22-depleted MCF7 cells with E2 and E2 plus 4-OHT) and those of SRC-3 were identified as a shared peak within 2 data sets (MCF7 cells with E2 and Fbxo22-depleted MCF7 cells with E2). (B) Tag counts of SRC-3 binding within the 410 shared SRC-3 peaks shown in A. ****P < 0.001, by 1-way ANOVA. (C) Heatmap visualization of ER and SRC-3 in the indicated MCF7 cells. (D) Genome browser snapshot of ChIP-Seq samples for ER (black) and SRC-3 (green) in GREB1 and IGFBP4 gene loci in the indicated MCF7 cells.
Fbxo22 is required for TAM-induced inhibition of breast cancer cell growth both in vitro and in vivo. Estrogen is required for MCF7 cell growth (39). Therefore, we examined whether Fbxo22 is a prerequisite for growth suppression of MCF7 cells by 4-OHT. A colony formation assay revealed that treatment with 4-OHT completely suppressed MCF7 cell proliferation in the presence of E2, whereas Fbxo22 depletion decreased the suppressive activity that was reversed by reintroduction of WT, but not ΔFbox, Fbxo22 (Figure 7A and Supplemental Figure 10A). We observed similar results in 2 other breast cancer cell lines, ZR75-1 and T47D, whose proliferation was also found to be dependent on estrogen stimulation (Supplemental Figure 10B). KDM4B depletion also resulted in a marked suppression of E2-induced MCF7 cell growth, independent of the presence of Fbxo22 (Figure 7B).
 Figure 7
Figure 7Fbxo22 is required for 4-OHT–mediated inhibition of breast cancer cell growth both in vitro and in vivo. (A) The indicated Dox-shRNA-MCF7 cells with or without doxycycline-inducible FLAG-Fbxo22 were starved of E2 in the presence of doxycycline (1 μg/ml) for 72 hours, treated with E2 (10 nM) in the presence or absence of 4-OHT (100 nM) for 6 hours, and then subjected to a quantitative colony formation assay. Representative images and results (mean ± SD) are from 3 independent experiments. (B) MCF7 cells expressing the indicated doxycycline-inducible shRNAs were treated as in A and subjected to a quantitative colony formation assay. Results are shown as the mean ± SD of 3 independent experiments. (C) Tumor growth of the control (n = 5) or Fbxo22-KO (n = 5) T47D cells transplanted into the mammary fat pads of NOD/Scid mice was measured over a 2-week period with E2 pellet supplementation and then over a 4-week period with TAM pellet supplementation. *P < 0.05, by 2-tailed Welch’s t test. (D) Mice as in C were sacrificed 6 weeks after transplantation. The tumors were then excised and weighed. *P < 0.05, by 2-tailed Welch’s t test. (E) Images of tumors excised from mice as in D are shown. The weight of each tumor is indicated below the images. (F) Paraffin-embedded tumor sections from 5 mice harboring WT or Fbxo22-KO T47D cells were subjected to immunohistochemical analyses using anti–Ki-67 antibodies and antibodies against cleaved caspase 3. Ki-67–positive and cleaved caspase 3–positive cells were counted, and their numbers were normalized to that of cell nuclei in each section. *P < 0.05 and ***P < 0.05, by2-tailed Student’s t test.
We next examined whether Fbxo22 is also required for the antagonistic activity of TAM in vivo. Control or Fbxo22-KO T47D breast cancer cells (3 × 106 cells) were xenotransplanted into the mammary fat pads of female NOD/Scid mice. Two weeks after the transplantation, the mice were transplanted with a TAM pellet, and tumor growth was analyzed. Before TAM treatment, we observed that both control and Fbxo22-KO cells formed tumors of comparable size in all transplanted mice, indicating that Fbxo22 deletion, per se, did not affect T47D growth in female NOD/Scid mice. Remarkably, the mice that received Fbxo22-KO cells after TAM treatment showed progressive tumor growth, whereas those that received control cells did not (Figure 7C). Consistent with this, the colony formation of Fbxo22-KO T47D cells was significantly enhanced, even in the presence of E2 and 4-OHT when compared with parental and WT T47D cells, and this enhancement was effectively reversed when WT Fbxo22 was reintroduced (Supplemental Figure 10C). Six weeks after transplantation, the mice were sacrificed, and the tumors were analyzed. We found that the tumors composed of Fbxo22-KO T47D cells were significantly heavier than those composed of WT T47D cells (Figure 7D). Representative images of pairs of tumors are shown in Figure 7E. Immunohistochemical analyses using tumor tissues revealed reduced apoptosis and increased proliferation of T47D cells lacking Fbxo22 (Figure 7F). Taken together, these results suggested that Fbxo22 is essential for the antagonistic activity of TAM in vitro and in vivo through selective degradation of KDM4B complexed with ER.
Fbxo22 expression predicts outcomes in patients with ER-positive/HER2-negative breast cancers. The essential role of SCFFbxo22 as a determining factor of TAM sensitivity in breast cancer cells suggests that the level of Fbxo22 may be used as a predictive value for patients with ER-positive breast cancers. To explore this possibility, we performed immunohistochemical analysis of a set of 163 primary ER-positive/HER2-negative breast cancer specimens to determine Fbxo22 levels. The immunohistochemical study revealed heterogeneous nuclear staining in healthy mammary glands (Figure 8A), whereas homogenous nuclear staining was detected in tumor tissue (Figure 8B, left panel). In some cases, staining was not detected (Figure 8B, right panel). We scored a specimen as negative for Fbxo22 staining when less than 1% of the tumor cells showed nuclear staining. Of the 163 specimens, 49 (30.1%) were scored as negative for Fbxo22 (Supplemental Table 1 and Supplemental Figure 11A). Fbxo22 negativity was correlated with high Ki-67 levels and a negative progesterone receptor (PR) status, a result possibly reflecting the aggressive biological features of the protein (Supplemental Table 1). However, Fbxo22 status was not correlated with lymph node involvement or tumor grade (Supplemental Table 1). Remarkably, the tumors lacking Fbxo22 expression were significantly associated with reduced relapse-free survival (RFS) compared with tumors positive for Fbxo22 (Figure 8C). This significant difference was preserved in separate cohorts of luminal A–like (low Ki-67), node-negative, and grade-1 tumor cases with even higher hazard ratios (HRs) (Figure 8, D and E, and Supplemental Figure 11B). Furthermore, Fbxo22-negative breast cancer showed significantly poorer RFS in a cohort of patients who received TAM treatment (Figure 8F), but not in a cohort of patients who were not given TAM (Supplemental Figure 11C). In the total cohort of 163 patients, the Ki-67–high, node-positive, and tumor grade-2/-3 patients had poorer outcomes, although the differences were not statistically significant (Supplemental Figure 11, D–F). In multivariate survival analyses, the lack of Fbxo22 was independently predictive of a poorer RFS (Supplemental Table 2). Together, the clinical data indicated that lack of Fbxo22 results in a poorer outcome in ER-positive/HER2-negative breast cancer, regardless of low Ki-67 expression, node-negative status, low tumor grade, or treatment with TAM. The association of Fbxo22 expression with RFS in ER-positive/HER2-negative breast cancer was further validated in another patient cohort at a different institution with a monoclonal antibody that we established (Figure 8G and see Supplemental Figures 16, A and B). Furthermore, we analyzed the association of Fbxo22 expression with RFS in ER-negative breast cancer. Low expression levels of Fbxo22 were not associated with poor outcomes, but rather with relatively better outcomes (Supplemental Figure 12), suggesting that the observed effect of Fbxo22 on breast cancer prognosis is specific to ER-positive cancers. These results may be attributed to Fbxo22 suppression causing resistance to TAM, as indicated by our in vitro results. Recently, a poor prognosis for breast cancer patients with low Fbxo22 expression levels was also reported, although the authors proposed an ER-unrelated function of Fbxo22 in breast cancer metastasis, and subset analysis for prognosis of the subtypes was not addressed (40).
 Figure 8
Figure 8Fbxo22 loss predicts a poorer outcome in patients with ER-positive/HER2-negative breast cancers. Representative images of normal mammary gland tissue (A) and positive (left) and negative (right) immunohistochemical staining for Fbxo22 in human breast cancer tissue (B). Scale bars: 20 μm. RFS was stratified by Fbxo22 protein expression in all T2 ER-positive/HER2-negative breast cancers (C); luminal A–like (low Ki-67) breast cancers (D); node-negative breast cancers (E); TAM-treated breast cancers (F); and in another cohort of T2 ER-positive/HER2-negative breast cancers stained with Fo-22, a monoclonal antibody established in the current study (clinicopathological variables are listed in Supplemental Table 3) (G). Kaplan-Meier survival curves are shown for patients whose cells were positive for Fbxo22 staining (red lines) and for patients whose cells were negative for Fbxo22 staining (blue lines). P values and HRs were calculated using a log-rank test.
-
Discussion
There is intense interest in understanding the molecular mechanisms underlying the action of SERMs, because this will potentially provide new multifunctional medicines that can be applied to the treatment of breast cancers, endometrial cancers, osteoporosis, and coronary heart disease. In the current study, we provide what we believe to be a new concept of SERM regulation, in which SCFFbxo22-mediated degradation of KDM4B is critical for the antagonistic activity of SERMs. These conclusions are supported by the following lines of evidence: (a) KDM4B depletion caused dissociation of SRC-3 from ER in the presence of E2. (b) SCFFbxo22 specifically ubiquitylated KDM4B complexed with unliganded or SERM-bound ER in vitro. (c) The real-time cellular dynamics in living cells revealed that Fbxo22 is essential for the release of SRC-1 in the presence of 4-OHT. (d) Genome-wide analyses showed that Fbxo22 is required for SRC-3 release by 4-OHT from almost all ER–SRC-3–bound genomic regions. (e) Either Fbxo22 or KDM4B degradation is essential for SERM-mediated suppression of ER-regulated transcription and breast cancer cell growth both in vitro and in vivo. Importantly, the agonistic activity of 4-OHT in the absence of Fbxo22 required AF1 activity. Recent structural analysis of the ER–SRC-3 complex on DNA revealed that AF1 located proximal to AF2 participates in SRC-3 recruitment in collaboration with AF2 (41). Therefore, we postulate that KDM4B degradation by SCFFbxo22 causes a conformational change that allows dissociation of SRC-3 from the AF1 domain, while TAM, per se, is sufficient for the release of SRC-3 from the AF2 domain. Taken together, our findings provide a common mechanism underlying the antagonistic activity of SERMs and show that the level of Fbxo22 or KDM4B may be a critical determinant of the selective modulation activities of SERMs (Supplemental Figure 13).
KDM4B has been reported to be essential for ER-mediated transcription, at least in part through demethylation of H3K9me3/me2 on the target promoters (26, 28, 29). Thus, KDM4B probably regulates ER signaling by both changing the chromatin structure on responsive promoters and acting as a scaffold for ER-coactivator recruitment. Indeed, the KDM family of proteins have been reported to have both catalytic and noncatalytic functions in gene regulation (23). We recently reported that SCFFbxo22 targeted p53 complexed with KDM4A for ubiquitylation, whereby the FIST-N domain of Fbxo22 binds to p53 and that of FIST-C binds to KDM4A. Although the binding of Fbxo22 to ER and KDM4B appeared to be independent (Figure 2G), both bindings drastically enhanced KDM4B ubiquitylation, suggesting that either binding is insufficient for substrate ubiquitylation. Thus, it is plausible that SCFFbxo22 preferentially targets transcription factor–KDM4 family lysine demethylase complexes and that the actual ubiquitylation target in the complex might be determined by entire structures or contexts.
Our current results also demonstrate that Fbxo22 status predicts the outcome in patients with ER-positive/HER2-negative breast cancer. Although there are various publicly available databases such as The Cancer Genome Atlas (TCGA), in which changes in transcriptomes are comprehensively analyzed in breast cancers, none of them showed that the levels of Fbxo22 transcripts are valuable for the prediction of outcome in patients with ER-positive/HER2-negative breast cancers (Supplemental Figure 14, A–D). Thus, immunohistochemical analysis of Fbxo22 protein in ER-positive/HER2-negative breast cancers is required for this purpose. ER-positive/HER2-negative breast cancer has been subclassified in the narrow sense as luminal A on the basis of Ki-67 status in order to optimize adjuvant therapy (42, 43). However, a substantial number of patients with low Ki-67 luminal A–like breast cancer experience relapse after TAM treatment (43), thus prompting efforts to find better predictive markers (44, 45). In our cohort, patients with high Ki-67 levels had poorer RFS rates, with a HR of 1.54, compared with patients with low Ki-67 levels, as evaluated by univariate analyses, and this finding was concordant with that of a previous report (43), although the results were not statistically significant. Interestingly, the HR decreased to 1.04 in an evaluation using multivariate analysis, whereas loss of Fbxo22 was coupled with a high HR of 2.65, with a significant P value. Loss of Fbxo22 was significantly associated with poorer RFS, even in the Ki-67–negative cohort, node-negative cohort, and low tumor grade cohort. The loss of Fbxo22 appears to be an intrinsic factor leading to TAM resistance, since Fbxo22 levels did not vary in the ER-positive/HER2-negative breast cancers either before or after neoadjuvant hormone therapy (Supplemental Figure 15A), and since Fbxo22 levels in TAM-sensitive MCF7 cells were comparable to those in TAM-resistant cells, which were established by long-term TAM treatment in TAM-sensitive cells (Supplemental Figure 15B). Therefore, we propose that, as a clinical tool, Fbxo22 status can help to define who is at high risk among patients with ER-positive/HER2-negative breast cancer, a subclass that is generally categorized as low risk. Together with the biological data described in this report, the risk can probably be ascribed to the agonistic activity of SERMs through a defective cofactor exchange under low Fbxo22 expression levels. In addition, we do not deny the possibility that Fbxo22 deficiency may also contribute to resistance to aromatase inhibitors to a lesser extent, because dissociation of coactivators from ER under E2-depleted conditions also required functional Fbxo22 and a proteasomal degradation of KDM4B (Figure 1D and Supplemental Figure 5, A and B). On that note, lack of Fbxo22 also revealed a tendency toward poorer outcomes in TAM-untreated, ER-positive/HER2-negative breast cancer patients, most of whom were treated with aromatase inhibitors (Supplemental Figure 11C).
Given that KDM4B also plays an important role in androgen signaling in prostate cancer (46), Fbxo22 status may have a good predictive value for patients with AR-positive prostate cancers. Finally, our results provide molecular clues for developing next-generation selective nuclear receptor modulators with activities that can be controlled depending on the cell and tissue contexts as well as the type of breast cancer.
-
Methods
Antibodies, shRNAs, and primers. Information on the shRNAs, antibodies, and primers used in this study is summarized in Supplemental Tables 5–7, respectively. A monoclonal antibody (Fo-22) specific to Fbxo22 was generated in mice with the keyhole limpet hemocyanin–conjugated peptide CNEVKDDDLFHSYTTIMALIHLGSSK as an antigen. Antibody specificity was validated by ELISA and immunohistochemistry (Supplemental Figure 16, A and B).
Cell culture. Cell cultures and treatments with various drugs were performed as described previously (47). MCF7 (ATCC), TAM-resistant MCF7(MCF7/TAMR-1, MilliporeSigma), T47D (ATCC), ZR75-1 (ATCC), U2OS (ATCC), U2OS-LacO-I-SceI-TetO (MilliporeSigma) (38), or 293T (ATCC) cells were cultured in DMEM (Invitrogen, Thermo Fisher Scientific) supplemented with 10% FBS. E2 (MilliporeSigma), cycloheximide (MilliporeSigma), 4-OHT (MilliporeSigma), fulvestrant (MilliporeSigma), toremifene (MilliporeSigma), and MG132 (MilliporeSigma) were used at concentrations of 10 nM, 50 μg/ml, 100 nM, 10 μg/ml, 100 nM, and 100 nM, respectively. For E2 depletion, cells were washed with PBS 3 times and then incubated with phenol red–free DMEM supplemented with 5% charcoal-stripped FBS for 72 hours.
Colony formation assay. Cells (5 × 102) were plated in 6-well plates and incubated for 2 weeks. Colonies were fixed with methanol/acetic acid (1:1) for 15 minutes, stained with 0.4% trypan blue (MilliporeSigma) in 20% ethanol in PBS for 15 minutes and counted.
Plasmid construction. Lentivirus-based shRNA constructs and Tet-on–inducible lentivirus constructs were generated as described previously (47). In brief, to generate lentivirus-based shRNA constructs, a 19-21 base shRNA-coding fragment with a 5′-ACGTGTGCTGTCCGT-3′ loop was introduced into pENTR4-H1 digested with AgeI/EcoRI. To insert the H1tetOx1-shRNA into the lentivirus vector, we mixed the resulting pENTR4-H1-shRNA vector and CS-RfA-ETBsd or CS-RfA-ETPuro vector with Gateway LR clonase (Invitrogen, Thermo Fisher Scientific). To construct Tet-on–inducible lentivirus constructs, the PCR-generated BamHI/NotI fragments containing cDNA for human and mouse Fbxo22-WT or the respective mutants and human KDM4B-WT were inserted into a pENTR-1A vector (Invitrogen, Thermo Fisher Scientific) containing the FLAG, HA, FLAG-HA epitope, or EGFP digested with BamHI/NotI. The resultant plasmid was mixed with CS-IV-TRE-RfA-UbC-Puro or CS-IV-TRE-RfA-UbC-Hygro vector and reacted with Gateway LR clonase to generate the lentiviral plasmid. pcDNA3-(HA-Ub)x6 containing 6 tandem repeats of HA-tagged ubiquitin was generated as described previously (48). pcDNA3-St2-KDM4B was generated with KDM4B cDNA subcloned into pcDNA3 with the following oligonucleotide corresponding to 2 Strep II epitopes with a linker peptide: 5′-TGGAGCCATCCTCAGTTCGAGAAAGGTGGCGGTTCTGGCGGAGGGTCGGGCGGCTCCGCCTGGAGTCACCCTCAGTTTGAGAAA-3′. pcDNA3-HA-KDM4B, CFP-LacER, and YFP-SRC-1 were provided by Kristian Helin (BRIC, University of Copenhagen, Copenhagen, Denmark) and Michael A. Mancini (Baylor College of Medicine, Houston, Texas, USA), respectively.
Virus generation and infection. Generation of lentiviruses and their infection of cells were performed as described previously (47). Lentiviruses expressing the respective shRNAs or genes were generated by cotransfection of 293T cells with pCMV-VSV-G-RSV-RevB, pCAG-HIVgp, and the respective CS-RfA-ETBsd, CS-RfA-ETPuro, CS-IV-TRE-RfA-UbC-Puro, CS-IV-TRE-RfA-UbC-Hygro, or CSII-CMV-MCS using the calcium phosphate coprecipitation method. Virus-infected cells were treated with 10 μg/ml blasticidin (Invitrogen, Thermo Fisher Scientific) and/or 2 μg/ml puromycin (MilliporeSigma) for 2 to 3 days. Doxycycline (MilliporeSigma) was added to the medium at a concentration of 1 μg/ml for inducible expression of the respective shRNAs or genes.
Immunoprecipitation and immunoblotting analyses. Immunoprecipitation and immunoblotting were performed as described previously (47). Cells were lysed in Tris-buffered saline containing NP-40 (TBSN) (20 mM Tris-Cl, pH 8.0, 150 mM NaCl, 0.5% NP-40, 5 mM EGTA, 1.5 mM EDTA, and 0.5 mM Na3VO4). The resulting lysates were clarified by centrifugation at 15,000 g for 20 minutes at 4°C before immunoprecipitation with antibodies. For immunoprecipitation against ER and KDM4B, nuclear extracts were prepared as reported previously (49). In brief, cell pellets were suspended in a 5× packed cell volume of hypotonic buffer A (10 mM HEPES-KOH, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mMDTT, and 0.5 mM PMSF) supplemented with a cocktail of protease inhibitors (Nakalai Tesque) and incubated on ice for 5 minutes. Cells were then centrifuged at 500 g for 5 minutes at 4°C, suspended in a 2 packed cell volume of buffer A and lysed by Dounce homogenization using a tight-fitting pestle. Nuclei were collected as a pellet by centrifugation at 4,000 g for 5 minutes at 4°C and extracted in an equal volume of buffer B (20 mM HEPES-KOH, pH 7.9, 600 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 0.5 mM DTT, and 0.5 mM PMSF) supplemented with a protease cocktail, and then mixed on a rotator at 4°C for 30 minutes. Nuclear extracts (supernatants) were recovered by centrifugation (16,000 g for 15 minutes at 4°C) and dialyzed using Slide-A-Lyzer Dialysis Cassettes (3,500-D protein molecular weight cutoff; Thermo Fisher Scientific) against buffer C (20 mM HEPES-KOH, pH 7.9, 100 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.5 mM DTT, and 0.5 mM PMSF). Dialyzed nuclear extracts were centrifuged (16,000 g for 30 minutes at 4°C) to eliminate residual precipitates. For whole lysates, cells were directly lysed with Laemmli buffer (2% SDS, 10% glycerol, 5% 2-mercaptoethanol, 0.002% bromophenol blue, and 62.5 mM Tris HCl at pH 6.8). The whole-lysate proteins (20–50 μg) were separated by SDS-PAGE, transferred onto a PVDF membrane (Immobilon-P; MilliporeSigma), and then subjected to immunoblotting with antibodies using an ECL detection system.
Quantitative RT-PCR. Quantitative real-time PCR (qRT-PCR) was performed as described previously (47). Total RNA was extracted using ISOGEN II (Wako) according to the manufacturer’s instructions. For qRT-PCR analysis, cDNAs were synthesized using a SuperScript II cDNA Synthesis Kit (Invitrogen, Thermo Fisher Scientific). Real-time PCR amplifications were performed in 96-well optical reaction plates with Power SYBR Green PCR Master Mix (Applied Biosystems). The relative expression values of each gene were determined by normalization to GAPDH expression for each sample.
Ubiquitination assay. For detection of in vivo ubiquitylated KDM4B, cells were transfected with the plasmids including 2× Strep II–tagged (WSHPQFEKGGGSGGGSGGSAWSHPQFEK) KDM4B, treated with 20 μM MG132 for 16 hours, and harvested 48 hours after transfection. Cells were lysed under denaturing conditions with 1% SDS-containing buffer and then centrifuged, and the supernatant was diluted as described previously (48, 50). Pulldown with 10 μl of 50% StrepTactin Resin (IBA Lifesciences) was performed according to the manufacturer’s instructions using wash buffer containing a high salt concentration (2 M NaCl, 50 mM Tris-HCl, pH 7.5, 0.5% Nonidet P-40, 150 mM NaCl, 50 mM NaF, 1 mM dithiothreitol). The resin was boiled in sample buffer and subjected to immunoblotting.
Fluorescence microscopy. U2OS-LacO-I-SceI-TetO cells were cultured on a glass-bottomed dish (Iwaki) placed on the stage of a BZ-9000 (Keyence) equipped with an environmental chamber (Keyence) that provided appropriate temperature, humidity, and CO2 control. Images were analyzed using BZ-9000 software.
CRISPR/Cas9-mediated gene KO. sgRNAs for human Fbxo22 were obtained in the form of oligonucleotides, which were annealed and cloned into the dual Cas9 and sgRNA expression vector pX330 (provided by Feng Zhang, MIT, Cambridge, Massachusetts, USA) with a BbsI site as reported previously (51). The plasmid (pX330-hFbxo22-4) was transfected into MCF7 or T47D cells using Lipofectamine 3000 (Invitrogen, Thermo Fisher Scientific) according to the manufacturer’s protocol. After a 48-hour incubation, the cells were split individually to generate a clonal cell line. Cell lysates of each cell line were subjected to Western blotting using anti-Fbxo22 antibody to confirm gene disruption. The sgRNA sequence for human Fbxo22 was 5′-CGCCGGAACCAGTCCTACGG-3′.
ChIP-Seq analysis. ChIP was performed with a SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology) according to the manufacturer’s instructions. Briefly, approximately 4 × 106 cells were fixed with 1% formaldehyde for 10 minutes at room temperature, followed by quenching with 125 mM glycine. Chromatin was prepared from cell pellets and digested with micrococcal nuclease for 15 minutes at room temperature. The digested chromatin was incubated with approximately 2 μg of the antibody at 4°C overnight, followed by incubation with 20 μl magnetic beads at 4°C for 2 hours. The beads were thoroughly washed 4 times with the wash buffer, and the chromatin was eluted with the ChIP elution buffer, followed by the reversal of cross-linking with protein K at 65°C for 4 hours. DNA was then purified on the DNA purification column. Sequencing libraries were prepared with 1.8 ng of the precipitated DNAs using an Ion Xpress Plus Fragment Library Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Sequencing was carried out with an Ion Proton system using an Ion PI Chip and Ion PI Sequencing 200 Kit (both from Thermo Fisher Scientific). Base calling for single-end reads and alignment were performed using Torrent Suite Software, version 5.2.2 (Thermo Fisher Scientific), with default settings. The reads were mapped to reference human genome hg19 using the Torrent Mapping Alignment Program (TMAP) provided with Torrent Suite. Tag directories were created with the aligned reads using the makeTagDirectory supplied in HOMER, version 4.9.1 (http://homer.salk.edu/homer/). We then performed basic quality control analyses and a sequence bias analysis using HOMER. Peaks were called using findPeaks in HOMER with the default parameters provided, namely a tag density of more than 4-fold and a FDR below 0.001, compared with the normalized tag density in the 10-kb region surrounding each peak. To find enriched TF motifs, we used findMotifsGenome (HOMER) with a region size of 50 bp and a P value less than 1 × 10–50 and annotated the regions using annotatePeaks (HOMER). Overlapping ERα and SRC-3 peaks were identified using a distance of less than 100 bp between the enriched regions of ERα and SRC-3. Hierarchical cluster analysis of the enriched regions was performed using Cluster 3.0. The heatmap data matrix was generated using HOMER and then visualized on Java TreeView (SourceForge). The box-and-whisker plots were created using a logarithm 2 of each density in Excel. All ChIP-Seq data for the shControl and shFbxo22 MCF7 cells in this study were deposited in the NCBI’s Gene Expression Omnibus (GEO) database (GEO GSE119702).
Mouse xenografts. Control or Fbxo22-KO T47D cells were grown to 80% to 90% confluence, trypsinized, resuspended in PBS, and mixed 1:1 with Matrigel (Corning, 354230). Estrogen pellets (60-day slow-release pellet containing 0.72 mg; Innovative Research of America) were implanted s.c. into the nape of the neck 1 day before 3 × 106 cells were injected s.c. with 100 μl of 1:1 PBS/Matrigel into the mammary fat pads of NOD/Scid mice (CLEA Japan Inc.) at 9 weeks after birth. When the tumor size reached approximately 50 mm3, five mice in each group were treated with TAM pellets (60-day slow-release pellet containing 5 mg ; Innovative Research of America) via s.c. implantation. Mice were measured weekly for tumor growth. After 6 weeks, the mice were euthanized and the tumors sectioned, measured, and then formalin fixed and paraffin embedded, and H&E-stained slides were prepared. Tumor sizes were estimated as follows: volume = (length × width × height × 0.5326) mm3. All animals were maintained under specific pathogen–free conditions.
Immunohistochemical analysis of tumor sections. Engrafted tumor tissues were fixed in 10% neutral-buffered formalin, dehydrated, and embedded in paraffin. Paraffin sections were deparaffinized, rehydrated, and incubated with antibodies against Ki-67 (M7240, Dako) or cleaved caspase-3 (5A1E, CST). Primary antibody binding was detected using an HRP-linked secondary antibody and visualized by incubation with DAB as a substrate.
Patients, tissue specimens, and immunohistochemistry. A panel of formalin-fixed, paraffin-embedded archival core-needle biopsies from 163 cases of consecutively treated primary T2 (2–5 cm in diameter) ER-positive/HER2-negative breast cancer and 41 cases of E2-negative breast cancer and the relevant clinical data were obtained from patients undergoing surgery at University Hospital, St. Marianna University School of Medicine, between 2005 and 2009. The median follow-up period was 7.4 and 9.5 years for the ER-positive/HER2-negative and ER-negative cohorts, respectively. The clinicopathological variables of the patients are listed in Supplemental Tables 1 and 4. For the validation cohort, core-needle biopsies (for patients who underwent neoadjuvant therapy) or surgical specimens from 30 cases of T2 ER-positive/HER2-negative breast cancer and the relevant clinical data were obtained from patients undergoing surgery at University Hospital, Hyogo College of Medicine, between 2009 and 2016. The median follow-up period was 3.9 years. Clinicopathological variables for the patients are listed in Supplemental Table 3.
Immunohistochemical analysis was performed with a Nichirei Histofine system (Nichirei Biosciences Inc.). Tissue sections were incubated with primary anti-Fbxo22 antibody (GTX117774, GeneTex, or monoclonal anti-Fbxo22 antibody Fo-22 for the validation cohort) at a 1:200 dilution and detected with HRP-labeled polymer-conjugated secondary antibody (Histofine Simple Stain MAX PO, multi, Nichirei). Color development was achieved with 3,3′-diaminobenzidine tetrahydrochloride. Cancer tissue with 1 or more cells showing moderate or strong nuclear Fbxo22 staining of 100 cells examined was judged as Fbxo22 positive. Positivity was evaluated in a blinded manner by 2 pathologists (IM and Yasushi Arizumi, St. Marianna University School of Medicine, Kawasaki, Japan) to reach a consensus for each case. The status of ER, PR, HER2, and Ki-67 was determined using standard immunohistochemical and FISH techniques for clinical analyses.
Statistics. Statistical analysis of cell-based experiments was performed using the 2-tailed Student’s t test for independent variables. P values of less than 0.05 were considered statistically significant. Statistical analysis of tag counts in ChIP-Seq data was performed using 1-way ANOVA. The relationship between Fbxo22 status and various clinicopathologic features was calculated with a χ2 test and Fisher’s exact test for categorical variables and a Student’s t test for continuous variables. RFS curves were constructed using the Kaplan-Meier method, and a log-rank test was used to evaluate differences in the survival curves. A Cox proportional hazards regression model was used to estimate the HR and 95% CIs of RFS for each variable in the univariate and multivariate analyses. HRs and 95% CIs with 2-sided P values are presented. The Kaplan-Meier plot was constructed using GraphPad Prism 6. Cox proportional hazards regression models were examined using R (version 3.3.2). Statistical significance was defined as a P value of less than 0.05.
Study approval. The clinical study conducted at the St. Marianna University School of Medicine was approved by the IRB of St. Marianna University (registration number 3095). This clinical study did not require informed consent, and patients were given the option to opt out. The validation study performed at the Hyogo College of Medicine was approved by the ethics committee of the Hyogo College of Medicine (registration number 106), and informed consent was obtained from all participants. Animal experiments were approved by the Animal Resource Center of The University of Tokyo (A16-33) and performed in accordance with IACUC guidelines and regulations.
-
Author contributions
MN and T. Ohta planned all experiments, analyzed data, and wrote the manuscript. YJ and WW performed the experiments and analyzed data. IM, AG, SN, YK, KT, NI, YM, T. Osako, FA, RM, and T. Ohta performed immunohistochemical analysis with human samples and analyzed data. NS, MY, and JII performed xenograft experiments and analyzed data. MM, KY, and YF performed ChIP-Seq and analyzed data.
-
Acknowledgments
We are grateful to K. Helin (Memorial Sloan Kettering Cancer Center, New York, New York, USA), M.A. Mancini (Baylor College of Medicine, Houston, Texas, USA), and H. Miyoshi (Keio University, Tokyo, Japan) for reagents. We thank A. Nishiyama (Institute of Medical Science, University of Tokyo, Tokyo, Japan) for discussions, Y. Chiba (Institute of Medical Science, University of Tokyo) and Y. Togashi (St. Marianna University Graduate School of Medicine) for technical support, and Y. Azumi (St. Marianna University Graduate School of Medicine) for evaluation of immunohistochemical staining. MN was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology/Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (MEXT/JSPS KAKENHI) (JP26250027, JP22118003, and JP16K15239); by grants from the Japan Agency for Medical Research and Development (AMED) (JP17cm0106122, JP17fk0310111, and JP17gm5010001); as well as by grants from the Ono Medical Research Foundation, the Princess Takamatsu Cancer Research Fund, and the Relay for Life Japan Cancer Society. YJ was supported by a grant from the MEXT/JSPS KAKENHI (16H06148). T. Ohta was supported by a grant from the MEXT/JSPS KAKENHI (JP26290042, JP 24112005, and 17H03585) and from the AMED (JP16ck0106085h0003). WW was supported by grants from the MEXT/JSPS KAKENHI (JP26460402 and 17K08676).
Address correspondence to: Makoto Nakanishi, Division of Cancer Cell Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. Phone: 81.3.5449.5241; Email: mkt-naka@ims.u-tokyo.ac.jp. Or to: Tomohiko Ohta, Department of Translational Oncology, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae-ku, Kawasaki 216-8511, Japan. Phone: 81.44.977.8111; Email: to@marianna-u.ac.jp.
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2018;128(12):5603–5619.https://doi.org/10.1172/JCI121679.
-
References
- Tora L, et al. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989;59(3):477–487.
- Kumar R, Thompson EB. The structure of the nuclear hormone receptors. Steroids. 1999;64(5):310–319.
- McKenna NJ, Xu J, Nawaz Z, Tsai SY, Tsai MJ, O’Malley BW. Nuclear receptor coactivators: multiple enzymes, multiple complexes, multiple functions. J Steroid Biochem Mol Biol. 1999;69(1-6):3–12.
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103(6):843–852.
- Belandia B, Orford RL, Hurst HC, Parker MG. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 2002;21(15):4094–4103.
- Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998;339(22):1609–1618.
- Coates AS, et al. Tailoring therapies--improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann Oncol. 2015;26(8):1533–1546.
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365(9472):1687–1717.
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG), et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378(9793):771–784.
- Davies C, et al. Long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years after diagnosis of oestrogen receptor-positive breast cancer: ATLAS, a randomised trial. Lancet. 2013;381(9869):805–816.
- Nettles KW, Greene GL. Ligand control of coregulator recruitment to nuclear receptors. Annu Rev Physiol. 2005;67:309–333.
- Smith CL, O’Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25(1):45–71.
- Jordan VC, Gapstur S, Morrow M. Selective estrogen receptor modulation and reduction in risk of breast cancer, osteoporosis, and coronary heart disease. J Natl Cancer Inst. 2001;93(19):1449–1457.
- Berry M, Metzger D, Chambon P. Role of the two activating domains of the oestrogen receptor in the cell-type and promoter-context dependent agonistic activity of the anti-oestrogen 4-hydroxytamoxifen. EMBO J. 1990;9(9):2811–2818.
- McInerney EM, Katzenellenbogen BS. Different regions in activation function-1 of the human estrogen receptor required for antiestrogen- and estradiol-dependent transcription activation. J Biol Chem. 1996;271(39):24172–24178.
- Tzukerman MT, et al. Human estrogen receptor transactivational capacity is determined by both cellular and promoter context and mediated by two functionally distinct intramolecular regions. Mol Endocrinol. 1994;8(1):21–30.View this article via: PubMed Google Scholar
- Shang Y, Brown M. Molecular determinants for the tissue specificity of SERMs. Science. 2002;295(5564):2465–2468.
- Smith CL, Nawaz Z, O’Malley BW. Coactivator and corepressor regulation of the agonist/antagonist activity of the mixed antiestrogen, 4-hydroxytamoxifen. Mol Endocrinol. 1997;11(6):657–666.
- Keeton EK, Brown M. Cell cycle progression stimulated by tamoxifen-bound estrogen receptor-alpha and promoter-specific effects in breast cancer cells deficient in N-CoR and SMRT. Mol Endocrinol. 2005;19(6):1543–1554.
- Girault I, et al. Expression analysis of estrogen receptor alpha coregulators in breast carcinoma: evidence that NCOR1 expression is predictive of the response to tamoxifen. Clin Cancer Res. 2003;9(4):1259–1266.View this article via: PubMed Google Scholar
- Osborne CK, et al. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J Natl Cancer Inst. 2003;95(5):353–361.
- Shou J, et al. Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J Natl Cancer Inst. 2004;96(12):926–935.
- Berry WL, Janknecht R. KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells. Cancer Res. 2013;73(10):2936–2942.
- Wissmann M, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9(3):347–353.
- Crea F, et al. The emerging role of histone lysine demethylases in prostate cancer. Mol Cancer. 2012;11:52. View this article via: PubMed Google Scholar
- Yang J, et al. The histone demethylase JMJD2B is regulated by estrogen receptor alpha and hypoxia, and is a key mediator of estrogen induced growth. Cancer Res. 2010;70(16):6456–6466.
- Shi L, et al. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc Natl Acad Sci U S A. 2011;108(18):7541–7546.
- Kawazu M, et al. Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS ONE. 2011;6(3):e17830.
- Gaughan L, et al. KDM4B is a master regulator of the estrogen receptor signalling cascade. Nucleic Acids Res. 2013;41(14):6892–6904.
- Vrba L, Junk DJ, Novak P, Futscher BW. p53 induces distinct epigenetic states at its direct target promoters. BMC Genomics. 2008;9:486. View this article via: PubMed Google Scholar
- Tan MK, Lim HJ, Harper JW. SCF(FBXO22) regulates histone H3 lysine 9 and 36 methylation levels by targeting histone demethylase KDM4A for ubiquitin-mediated proteasomal degradation. Mol Cell Biol. 2011;31(18):3687–3699.
- Johmura Y, et al. SCF(Fbxo22)-KDM4A targets methylated p53 for degradation and regulates senescence. Nat Commun. 2016;7:10574. View this article via: PubMed Google Scholar
- Stenoien DL, et al. FRAP reveals that mobility of oestrogen receptor-alpha is ligand- and proteasome-dependent. Nat Cell Biol. 2001;3(1):15–23.
- Reid G, et al. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol Cell. 2003;11(3):695–707.
- Xu J, Li Q. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol Endocrinol. 2003;17(9):1681–1692.
- Lahusen T, Henke RT, Kagan BL, Wellstein A, Riegel AT. The role and regulation of the nuclear receptor co-activator AIB1 in breast cancer. Breast Cancer Res Treat. 2009;116(2):225–237.
- Stenoien DL, et al. Ligand-mediated assembly and real-time cellular dynamics of estrogen receptor alpha-coactivator complexes in living cells. Mol Cell Biol. 2001;21(13):4404–4412.
- Burgess RC, Burman B, Kruhlak MJ, Misteli T. Activation of DNA damage response signaling by condensed chromatin. Cell Rep. 2014;9(5):1703–1717.
- Ciocca DR, Fanelli MA. Estrogen receptors and cell proliferation in breast cancer. Trends Endocrinol Metab. 1997;8(8):313–321.
- Sun R, et al. FBXO22 Possesses Both Protumorigenic and Antimetastatic Roles in Breast Cancer Progression. Cancer Res. 2018;78(18):5274–5286.
- Yi P, et al. Structure of a biologically active estrogen receptor-coactivator complex on DNA. Mol Cell. 2015;57(6):1047–1058.
- Goldhirsch A, et al. Strategies for subtypes--dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22(8):1736–1747.
- Cheang MC, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101(10):736–750.
- Prat A, et al. Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal A breast cancer. J Clin Oncol. 2013;31(2):203–209.
- Azim HA, et al. Utility of prognostic genomic tests in breast cancer practice: The IMPAKT 2012 Working Group Consensus Statement. Ann Oncol. 2013;24(3):647–654.
- Coffey K, et al. The lysine demethylase, KDM4B, is a key molecule in androgen receptor signalling and turnover. Nucleic Acids Res. 2013;41(8):4433–4446.
- Johmura Y, et al. Necessary and sufficient role for a mitosis skip in senescence induction. Mol Cell. 2014;55(1):73–84.
- Nishikawa H, et al. Mass spectrometric and mutational analyses reveal Lys-6-linked polyubiquitin chains catalyzed by BRCA1-BARD1 ubiquitin ligase. J Biol Chem. 2004;279(6):3916–3924.
- Hirokawa T, et al. CBP-93872 inhibits NBS1-mediated ATR activation, abrogating maintenance of the DNA double-strand break-specific G2 checkpoint. Cancer Res. 2014;74(14):3880–3889.
- Sato K, et al. Nucleophosmin/B23 is a candidate substrate for the BRCA1-BARD1 ubiquitin ligase. J Biol Chem. 2004;279(30):30919–30922.
- Shimada M, et al. Essential role of autoactivation circuitry on Aurora B-mediated H2AX-pS121 in mitosis. Nat Commun. 2016;7:12059. View this article via: PubMed Google Scholar
-
Version history
- Version 1 (November 12, 2018): Electronic publication
- Version 2 (December 3, 2018): Print issue publication



Copyright © 2026 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)








