Advertisement
Research ArticleNeuroscience Free access | 10.1172/JCI93924
Cografting astrocytes improves cell therapeutic outcomes in a Parkinson’s disease model
Jae-Jin Song,1,2,3 Sang-Min Oh,1,2,3 Oh-Chan Kwon,1,2,3 Noviana Wulansari,1,2,3 Hyun-Seob Lee,4,5 Mi-Yoon Chang,1,2 Eunsoo Lee,6 Woong Sun,6 Sang-Eun Lee,7 Sunghoe Chang,7 Heeyoung An,8,9,10 C. Justin Lee,8,9,10 and Sang-Hun Lee1,2,3
1Department of Biochemistry and Molecular Biology, College of Medicine,
2Hanyang Biomedical Research Institute, and
3Graduate School of Biomedical Science and Engineering, Hanyang University, Seoul, South Korea.
4Genomic Core Facility, Transdisciplinary Research and Collaboration Division, Translational Research Institute, and
5Biomedical Research Institute, Seoul National University Hospital, Seoul, South Korea.
6Department of Anatomy and Division of Brain Korea 21 PLUS Program for Biomedical Science, Korea University College of Medicine, Seoul, South Korea.
7Department of Physiology and Biomedical Sciences, Seoul National University College of Medicine, Seoul, South Korea.
8KU-KIST Graduate School of Converging Science and Technology, Korea University, Seoul, South Korea.
9Center for Neuroscience and
10Center for Glia-Neuron Interaction, Korea Institute of Science and Technology (KIST), Seoul, South Korea.
Address correspondence to: Sang-Hun Lee, Department of Biochemistry and Molecular Biology, College of Medicine, Hanyang University, 17 Haengdang-dong, Sungdong-gu, Seoul 133-791, South Korea. Phone: 82.2.2220.0625; Email: leesh@hanyang.ac.kr.
Find articles by Song, J. in: PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology, College of Medicine,
2Hanyang Biomedical Research Institute, and
3Graduate School of Biomedical Science and Engineering, Hanyang University, Seoul, South Korea.
4Genomic Core Facility, Transdisciplinary Research and Collaboration Division, Translational Research Institute, and
5Biomedical Research Institute, Seoul National University Hospital, Seoul, South Korea.
6Department of Anatomy and Division of Brain Korea 21 PLUS Program for Biomedical Science, Korea University College of Medicine, Seoul, South Korea.
7Department of Physiology and Biomedical Sciences, Seoul National University College of Medicine, Seoul, South Korea.
8KU-KIST Graduate School of Converging Science and Technology, Korea University, Seoul, South Korea.
9Center for Neuroscience and
10Center for Glia-Neuron Interaction, Korea Institute of Science and Technology (KIST), Seoul, South Korea.
Address correspondence to: Sang-Hun Lee, Department of Biochemistry and Molecular Biology, College of Medicine, Hanyang University, 17 Haengdang-dong, Sungdong-gu, Seoul 133-791, South Korea. Phone: 82.2.2220.0625; Email: leesh@hanyang.ac.kr.
Find articles by Oh, S. in: PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology, College of Medicine,
2Hanyang Biomedical Research Institute, and
3Graduate School of Biomedical Science and Engineering, Hanyang University, Seoul, South Korea.
4Genomic Core Facility, Transdisciplinary Research and Collaboration Division, Translational Research Institute, and
5Biomedical Research Institute, Seoul National University Hospital, Seoul, South Korea.
6Department of Anatomy and Division of Brain Korea 21 PLUS Program for Biomedical Science, Korea University College of Medicine, Seoul, South Korea.
7Department of Physiology and Biomedical Sciences, Seoul National University College of Medicine, Seoul, South Korea.
8KU-KIST Graduate School of Converging Science and Technology, Korea University, Seoul, South Korea.
9Center for Neuroscience and
10Center for Glia-Neuron Interaction, Korea Institute of Science and Technology (KIST), Seoul, South Korea.
Address correspondence to: Sang-Hun Lee, Department of Biochemistry and Molecular Biology, College of Medicine, Hanyang University, 17 Haengdang-dong, Sungdong-gu, Seoul 133-791, South Korea. Phone: 82.2.2220.0625; Email: leesh@hanyang.ac.kr.
Find articles by Kwon, O. in: PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology, College of Medicine,
2Hanyang Biomedical Research Institute, and
3Graduate School of Biomedical Science and Engineering, Hanyang University, Seoul, South Korea.
4Genomic Core Facility, Transdisciplinary Research and Collaboration Division, Translational Research Institute, and
5Biomedical Research Institute, Seoul National University Hospital, Seoul, South Korea.
6Department of Anatomy and Division of Brain Korea 21 PLUS Program for Biomedical Science, Korea University College of Medicine, Seoul, South Korea.
7Department of Physiology and Biomedical Sciences, Seoul National University College of Medicine, Seoul, South Korea.
8KU-KIST Graduate School of Converging Science and Technology, Korea University, Seoul, South Korea.
9Center for Neuroscience and
10Center for Glia-Neuron Interaction, Korea Institute of Science and Technology (KIST), Seoul, South Korea.
Address correspondence to: Sang-Hun Lee, Department of Biochemistry and Molecular Biology, College of Medicine, Hanyang University, 17 Haengdang-dong, Sungdong-gu, Seoul 133-791, South Korea. Phone: 82.2.2220.0625; Email: leesh@hanyang.ac.kr.
Find articles by Wulansari, N. in: PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology, College of Medicine,
2Hanyang Biomedical Research Institute, and
3Graduate School of Biomedical Science and Engineering, Hanyang University, Seoul, South Korea.
4Genomic Core Facility, Transdisciplinary Research and Collaboration Division, Translational Research Institute, and
5Biomedical Research Institute, Seoul National University Hospital, Seoul, South Korea.
6Department of Anatomy and Division of Brain Korea 21 PLUS Program for Biomedical Science, Korea University College of Medicine, Seoul, South Korea.
7Department of Physiology and Biomedical Sciences, Seoul National University College of Medicine, Seoul, South Korea.
8KU-KIST Graduate School of Converging Science and Technology, Korea University, Seoul, South Korea.
9Center for Neuroscience and
10Center for Glia-Neuron Interaction, Korea Institute of Science and Technology (KIST), Seoul, South Korea.
Address correspondence to: Sang-Hun Lee, Department of Biochemistry and Molecular Biology, College of Medicine, Hanyang University, 17 Haengdang-dong, Sungdong-gu, Seoul 133-791, South Korea. Phone: 82.2.2220.0625; Email: leesh@hanyang.ac.kr.
Find articles by Lee, H. in: PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology, College of Medicine,
2Hanyang Biomedical Research Institute, and
3Graduate School of Biomedical Science and Engineering, Hanyang University, Seoul, South Korea.
4Genomic Core Facility, Transdisciplinary Research and Collaboration Division, Translational Research Institute, and
5Biomedical Research Institute, Seoul National University Hospital, Seoul, South Korea.
6Department of Anatomy and Division of Brain Korea 21 PLUS Program for Biomedical Science, Korea University College of Medicine, Seoul, South Korea.
7Department of Physiology and Biomedical Sciences, Seoul National University College of Medicine, Seoul, South Korea.
8KU-KIST Graduate School of Converging Science and Technology, Korea University, Seoul, South Korea.
9Center for Neuroscience and
10Center for Glia-Neuron Interaction, Korea Institute of Science and Technology (KIST), Seoul, South Korea.
Address correspondence to: Sang-Hun Lee, Department of Biochemistry and Molecular Biology, College of Medicine, Hanyang University, 17 Haengdang-dong, Sungdong-gu, Seoul 133-791, South Korea. Phone: 82.2.2220.0625; Email: leesh@hanyang.ac.kr.
Find articles by Chang, M. in: PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology, College of Medicine,
2Hanyang Biomedical Research Institute, and
3Graduate School of Biomedical Science and Engineering, Hanyang University, Seoul, South Korea.
4Genomic Core Facility, Transdisciplinary Research and Collaboration Division, Translational Research Institute, and
5Biomedical Research Institute, Seoul National University Hospital, Seoul, South Korea.
6Department of Anatomy and Division of Brain Korea 21 PLUS Program for Biomedical Science, Korea University College of Medicine, Seoul, South Korea.
7Department of Physiology and Biomedical Sciences, Seoul National University College of Medicine, Seoul, South Korea.
8KU-KIST Graduate School of Converging Science and Technology, Korea University, Seoul, South Korea.
9Center for Neuroscience and
10Center for Glia-Neuron Interaction, Korea Institute of Science and Technology (KIST), Seoul, South Korea.
Address correspondence to: Sang-Hun Lee, Department of Biochemistry and Molecular Biology, College of Medicine, Hanyang University, 17 Haengdang-dong, Sungdong-gu, Seoul 133-791, South Korea. Phone: 82.2.2220.0625; Email: leesh@hanyang.ac.kr.
Find articles by Lee, E. in: PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology, College of Medicine,
2Hanyang Biomedical Research Institute, and
3Graduate School of Biomedical Science and Engineering, Hanyang University, Seoul, South Korea.
4Genomic Core Facility, Transdisciplinary Research and Collaboration Division, Translational Research Institute, and
5Biomedical Research Institute, Seoul National University Hospital, Seoul, South Korea.
6Department of Anatomy and Division of Brain Korea 21 PLUS Program for Biomedical Science, Korea University College of Medicine, Seoul, South Korea.
7Department of Physiology and Biomedical Sciences, Seoul National University College of Medicine, Seoul, South Korea.
8KU-KIST Graduate School of Converging Science and Technology, Korea University, Seoul, South Korea.
9Center for Neuroscience and
10Center for Glia-Neuron Interaction, Korea Institute of Science and Technology (KIST), Seoul, South Korea.
Address correspondence to: Sang-Hun Lee, Department of Biochemistry and Molecular Biology, College of Medicine, Hanyang University, 17 Haengdang-dong, Sungdong-gu, Seoul 133-791, South Korea. Phone: 82.2.2220.0625; Email: leesh@hanyang.ac.kr.
Find articles by Sun, W. in: PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology, College of Medicine,
2Hanyang Biomedical Research Institute, and
3Graduate School of Biomedical Science and Engineering, Hanyang University, Seoul, South Korea.
4Genomic Core Facility, Transdisciplinary Research and Collaboration Division, Translational Research Institute, and
5Biomedical Research Institute, Seoul National University Hospital, Seoul, South Korea.
6Department of Anatomy and Division of Brain Korea 21 PLUS Program for Biomedical Science, Korea University College of Medicine, Seoul, South Korea.
7Department of Physiology and Biomedical Sciences, Seoul National University College of Medicine, Seoul, South Korea.
8KU-KIST Graduate School of Converging Science and Technology, Korea University, Seoul, South Korea.
9Center for Neuroscience and
10Center for Glia-Neuron Interaction, Korea Institute of Science and Technology (KIST), Seoul, South Korea.
Address correspondence to: Sang-Hun Lee, Department of Biochemistry and Molecular Biology, College of Medicine, Hanyang University, 17 Haengdang-dong, Sungdong-gu, Seoul 133-791, South Korea. Phone: 82.2.2220.0625; Email: leesh@hanyang.ac.kr.
Find articles by Lee, S. in: PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology, College of Medicine,
2Hanyang Biomedical Research Institute, and
3Graduate School of Biomedical Science and Engineering, Hanyang University, Seoul, South Korea.
4Genomic Core Facility, Transdisciplinary Research and Collaboration Division, Translational Research Institute, and
5Biomedical Research Institute, Seoul National University Hospital, Seoul, South Korea.
6Department of Anatomy and Division of Brain Korea 21 PLUS Program for Biomedical Science, Korea University College of Medicine, Seoul, South Korea.
7Department of Physiology and Biomedical Sciences, Seoul National University College of Medicine, Seoul, South Korea.
8KU-KIST Graduate School of Converging Science and Technology, Korea University, Seoul, South Korea.
9Center for Neuroscience and
10Center for Glia-Neuron Interaction, Korea Institute of Science and Technology (KIST), Seoul, South Korea.
Address correspondence to: Sang-Hun Lee, Department of Biochemistry and Molecular Biology, College of Medicine, Hanyang University, 17 Haengdang-dong, Sungdong-gu, Seoul 133-791, South Korea. Phone: 82.2.2220.0625; Email: leesh@hanyang.ac.kr.
Find articles by
Chang, S.
in:
PubMed
|
Google Scholar
|

1Department of Biochemistry and Molecular Biology, College of Medicine,
2Hanyang Biomedical Research Institute, and
3Graduate School of Biomedical Science and Engineering, Hanyang University, Seoul, South Korea.
4Genomic Core Facility, Transdisciplinary Research and Collaboration Division, Translational Research Institute, and
5Biomedical Research Institute, Seoul National University Hospital, Seoul, South Korea.
6Department of Anatomy and Division of Brain Korea 21 PLUS Program for Biomedical Science, Korea University College of Medicine, Seoul, South Korea.
7Department of Physiology and Biomedical Sciences, Seoul National University College of Medicine, Seoul, South Korea.
8KU-KIST Graduate School of Converging Science and Technology, Korea University, Seoul, South Korea.
9Center for Neuroscience and
10Center for Glia-Neuron Interaction, Korea Institute of Science and Technology (KIST), Seoul, South Korea.
Address correspondence to: Sang-Hun Lee, Department of Biochemistry and Molecular Biology, College of Medicine, Hanyang University, 17 Haengdang-dong, Sungdong-gu, Seoul 133-791, South Korea. Phone: 82.2.2220.0625; Email: leesh@hanyang.ac.kr.
Find articles by An, H. in: PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology, College of Medicine,
2Hanyang Biomedical Research Institute, and
3Graduate School of Biomedical Science and Engineering, Hanyang University, Seoul, South Korea.
4Genomic Core Facility, Transdisciplinary Research and Collaboration Division, Translational Research Institute, and
5Biomedical Research Institute, Seoul National University Hospital, Seoul, South Korea.
6Department of Anatomy and Division of Brain Korea 21 PLUS Program for Biomedical Science, Korea University College of Medicine, Seoul, South Korea.
7Department of Physiology and Biomedical Sciences, Seoul National University College of Medicine, Seoul, South Korea.
8KU-KIST Graduate School of Converging Science and Technology, Korea University, Seoul, South Korea.
9Center for Neuroscience and
10Center for Glia-Neuron Interaction, Korea Institute of Science and Technology (KIST), Seoul, South Korea.
Address correspondence to: Sang-Hun Lee, Department of Biochemistry and Molecular Biology, College of Medicine, Hanyang University, 17 Haengdang-dong, Sungdong-gu, Seoul 133-791, South Korea. Phone: 82.2.2220.0625; Email: leesh@hanyang.ac.kr.
Find articles by Lee, C. in: PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology, College of Medicine,
2Hanyang Biomedical Research Institute, and
3Graduate School of Biomedical Science and Engineering, Hanyang University, Seoul, South Korea.
4Genomic Core Facility, Transdisciplinary Research and Collaboration Division, Translational Research Institute, and
5Biomedical Research Institute, Seoul National University Hospital, Seoul, South Korea.
6Department of Anatomy and Division of Brain Korea 21 PLUS Program for Biomedical Science, Korea University College of Medicine, Seoul, South Korea.
7Department of Physiology and Biomedical Sciences, Seoul National University College of Medicine, Seoul, South Korea.
8KU-KIST Graduate School of Converging Science and Technology, Korea University, Seoul, South Korea.
9Center for Neuroscience and
10Center for Glia-Neuron Interaction, Korea Institute of Science and Technology (KIST), Seoul, South Korea.
Address correspondence to: Sang-Hun Lee, Department of Biochemistry and Molecular Biology, College of Medicine, Hanyang University, 17 Haengdang-dong, Sungdong-gu, Seoul 133-791, South Korea. Phone: 82.2.2220.0625; Email: leesh@hanyang.ac.kr.
Find articles by Lee, S. in: PubMed | Google Scholar
Published December 11, 2017 - More info
J Clin Invest. 2018;128(1):e93924. https://doi.org/10.1172/JCI93924.
© 2018 American Society for Clinical Investigation
Received: March 10, 2017; Accepted: October 31, 2017
Related article:
Abstract
Most of the adult CNS lacks regenerative activity in terms of both neuron birth and neurite outgrowth. While this regeneration-unfriendly environment of the adult CNS may preserve the existing neuronal circuitry that takes years to develop in higher organisms, it also poses a major obstacle for CNS repair later in life. In this issue of the JCI, Song et al. report on their development of a strategy that uses region-specific and molecularly engineered astrocytes to turn an unfavorable brain environment into a favorable one for engrafted neural stem/progenitor cells (NSC/NPCs). In a rat model of Parkinson’s disease (PD), cografting NPCs with midbrain-derived astrocytes engineered to overexpress the transcription factors Nurr1 and Foxa2 promotes maturation and survival of the graft, resulting in therapeutic improvement. The results of this study raise the prospect of using modified astrocytes to improve the survival, maturation, and integration of engrafted NSC/NPCs as a restorative treatment for PD.
Authors
Robert Y.L. Tsai
-
Abstract
Transplantation of neural progenitor cells (NPCs) is a potential therapy for treating neurodegenerative disorders, but this approach has faced many challenges and limited success, primarily because of inhospitable host brain environments that interfere with enriched neuron engraftment and function. Astrocytes play neurotrophic roles in the developing and adult brain, making them potential candidates for helping with modification of hostile brain environments. In this study, we examined whether astrocytic function could be utilized to overcome the current limitations of cell-based therapies in a murine model of Parkinson’s disease (PD) that is characterized by dopamine (DA) neuron degeneration in the midbrain. We show here that cografting astrocytes, especially those derived from the midbrain, remarkably enhanced NPC-based cell therapeutic outcomes along with robust DA neuron engraftment in PD rats for at least 6 months after transplantation. We further show that engineering of donor astrocytes with Nurr1 and Foxa2, transcription factors that were recently reported to polarize harmful immunogenic glia into the neuroprotective form, further promoted the neurotrophic actions of grafted astrocytes in the cell therapeutic approach. Collectively, these findings suggest that cografting astrocytes could be a potential strategy for successful cell therapeutic outcomes in neurodegenerative disorders.
-
Introduction
Given the clinical experience of fetal mesencephalic transplantation in Parkinson’s disease (PD), a neurodegenerative disorder characterized by dopamine (DA) neuron loss in the midbrain substantial nigra (SN), stem cell transplantation is regarded as a potential future therapy for treating intractable brain disorders. Neural progenitor cells (NPCs) can be derived from brain tissues or by in vitro differentiation of embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs). Therapeutic potentials for cultured NPCs have been shown in both in vitro and in vivo disease models. However, the general consensus is that therapeutic outcomes attained by current NPC transplantation techniques do not reach a level that is satisfactory for the treatment of patients (reviewed in ref. 1, 2). To overcome this limitation, studies have mainly centered on the improvement of intrinsic donor cell qualities. What is ignored, however, is that the host brain becomes hostile to grafted cells after transplantation due to immunogenic and inflammatory reactions induced by mechanical injury occurring during cell transplantation (3, 4), which hampers appropriate differentiation, maturation, survival, and function of the grafted cells (5). Thus, in vitro success in donor NPC cultures efficiently yielding authentic midbrain-type DA (mDA) neurons with improved survival and functions does not guarantee successful therapeutic outcomes after transplantation without correcting hostile brain environments.
Astrocytes, which outnumber neurons in the CNS, exert physiologic functions to support neuronal cell survival, neuronal function, and brain homeostasis in the adult brain as well as neuronal differentiation and synaptic maturation during development (6–8). The idea of modifying pathologic brain environments by utilizing the neurotrophic properties of astrocytes has been appearing on the therapeutic horizon for CNS disorders (9). However, multiple properties of astrocytes are compromised in various diseases (10, 11), and astrocytes can be activated into the type of cells establishing harmful and hostile brain environments in diseased contexts (12, 13) (frequently annotated as M1 activation using the terminology of macrophage polarization, but the terminology has not been completely established by research findings in microglia and astrocytes) (14). A previous study demonstrated that nuclear receptor–related factor 1 (Nurr1, also known as NR4A2), originally known as a transcription factor specific for developing and adult mDA neurons, could also be expressed in astrocytes/microglia in response to toxic insults and that Nurr1-expressing glia protect neighboring mDA neurons by reducing synthesis and releasing of proinflammatory cytokines from glial cells (15). In a later study, we showed that forkhead box protein A2 (Foxa2, also known as HNF3β) is a potent cofactor that synergizes the Nurr1-mediated antiinflammatory roles in glia (16). Furthermore, forced coexpression of Nurr1 and Foxa2 (Nurr1+Foxa2) in glia promotes the synthesis and release of various neurotrophic factors, indicating that engineering of Nurr1 and Foxa2 in glia could become a powerful strategy for treating CNS disorders.
In this study, we have attempted to exploit the neurotrophic actions of astrocytes and Nurr1+Foxa2 functions in this cell type to improve the therapeutic outcomes of NPC transplantation using an animal model of PD. Our in vitro assays using a coculture system and conditioned media treatment revealed that astrocytes, especially those cultured from the ventral midbrain (VM), where mDA neurons are developed and reside (hereafter referred as dopaminergic), greatly support a series of NPC behaviors associated with their therapeutic capacity upon transplantation, such as mDA neuron differentiation, synaptic maturation, midbrain-specific marker expression, presynaptic DA neuron function, and resistance against toxic stimuli. Nurr1+Foxa2 engineering in astrocytes further improved astrocytic function to protect mDA neurons against toxins, mainly by reducing inflammation. We further identified potential neurotrophic cytokines, extracellular matrix (ECM) proteins, and antiinflammatory and antioxidant factors that may mediate the actions of astrocytes. Based on these findings, we demonstrate the functional benefits of astrocyte cotransplantation in NPC-based cell therapy for PD.
-
Results
Immature and brain region–specific identities of astrocytes cultured from the cortex and VM of mouse pups. The aim of this study is to improve NPC transplantation efficacy by utilizing neurotrophic actions of astrocytes. Based on brain region–dependent diversity of astrocytes and their potential region-specific roles (17–21), we cultured astrocytes from 2 brain regions, including the classical nondopaminergic brain region cortex (Ctx) and dopaminergic VM. Along with high mRNA expression of astrocytic markers (Supplemental Figure 1B; supplemental material available online with this article; https://doi.org/10.1172/JCI93924DS1), immunocytochemical analyses revealed that major cell populations in the Ctx- and VM-astroglial cultures at day in vitro (DIV) 14 were positive for GFAP (85% and 82%), CD44 (79% and 76 %), GLT-1 (58% and 71 %), and GLAST protein (71% and 77%), respectively (Supplemental Figure 1A), except S100β (20% and 32%), a soluble protein associated with harmful reactivation of astrocytes (22). A few (0.24% ± 0.35% in Ctx-Ast and 0.43% ± 0.39% in VM-Ast, where Ast indicates astrocytes) and none of the cells in these cultures were Iba1+ microglia and O4+ oligodendrocyte, respectively. In contrast, none or a few cells were positive for the astroglial markers in the NPC cultures derived from embryonic cortices (Ctx-NPCs) used as a control (Supplemental Figure 1A). Along with abundant cells immunoreactive for the immature astrocytic markers GLAST and Sox2, the expression levels of the immature astrocytic genes (Sox2, Nestin, GLAST, and vimentin) in the RNA-sequencing (RNA-seq) data were high (fragments per kilobase of exons per million reads [FPKM]: 94–4180, Supplemental Figure 1C), indicating that the astrocytes cultured in this study had immature properties. Previous studies have reported that astrocytes in culture were electrophysiologically immature compared with those in vivo (23, 24). Consistently, in patch-clamp analyses, astrocytes cultured from cortices in this study (DIV14) exhibited current (2577 ± 401.1 pA, n = 16) and conductance (23.08 ± 3.306 nS, n = 16) that were significantly lower than those detected in cortical slices in vivo (40 nS) (23, 24). Interestingly, the current and conductance values of astrocytes cultured from VM at DIV14 were even lower compared with the cultured Ctx-Ast at DIV14, and those of VM-Ast increased after a longer culture period (at DIV35) (Supplemental Figure 1, D and E). Each group of astrocytes had a population with conductance lower than 10 nS. The percentage of low conductance cells was the highest in VM-Ast cultures at DIV14 and decreased during maturation in VM at DIV35, while the percentages were similar in Ctx DIV14 and Ctx DIV35 (Supplemental Figure 1F). Of note, it has been shown that transplanted immature — but not mature — astrocytes exert neurotrophic support in the injured CNS (25–29). In addition to a wound-healing capacity superior to that in astrocytes at DIV33–35 (Supplemental Figure 1G) in a scratch injury model (28), astrocytes at DIV12-14 showed greater capacity growing on high levels of chondroitin sulfate proteoglycans (CSPGs) (Supplemental Figure 1H) in an in vitro model recreating the strongly inhibitory CSPG gradient observed in the glial scar in vivo (25). These findings collectively indicated that cultured astrocytes in this study, especially those at DIV12–14, were immature and capable of exerting regenerative capacities. Based on these findings, the following experiments were done using astrocyte cultures at DIV7–14, unless otherwise noted.
Positional identity, a fundamental organizing principle governing the generation of neuronal subtype diversity, is relevant to astrocyte diversification (18, 21). Consistently, Ctx-Ast had high levels of expression of cortical region–specific genes (Emx2, Lhx2, FoxG1, and Pax6), whereas expression of the midbrain-specific markers (Foxa1/2, Lmx1a/b, and En1/2) was enriched in VM-Ast cultures (Supplemental Figure 1, I and J). While endogenous expression of Foxa2 was exclusively enriched in VM-Ast, expression of Nurr1, another major gene of interest in this study, and its downstream target gene Pitx3 (30) was indistinguishable between Ctx- and VM-Ast cultures (Supplemental Figure 1, I and J). This finding is consistent with the detection of Nurr1 expression not only in the midbrain, but also in cortical layer VI (31) and the hippocampus (32). These findings collectively verify immature astrocytic- and brain region–specific identities of the astrocyte cultures used in this study.
VM-Ast promote the differentiation of NPCs into mDA neurons with enhanced neuronal maturity, expression of midbrain-specific markers, and resistance against a toxic stimulus. In order to attain therapeutic efficacy by NPC transplantation in PD, grafted NPCs must undergo differentiation into mDA neurons, neuronal maturation with synapse formation, and survival of the grafted mDA neurons in the hostile environment of the grafted brain. We first assessed this series of the cellular events in the absence or presence of Ctx- or VM-Ast in vitro. To this end, NPCs isolated from rodent embryonic VMs (mouse at E10.5 or rat at E12) were expanded in vitro for 4 days, harvested, and mixed with the Ctx-Ast or VM-Ast at a cell number ratio of 2:1 VM-NPCs to astrocytes. The mixed cells were plated, and differentiation of VM-NPCs was directly induced in serum-free N2 medium (schematic in Figure 1A). Since cell differentiation is affected by cell density (33, 34), VM-NPCs in the control cultures were mixed with nondopaminergic Ctx-NPCs and we determined that cell density (confluence) among the tested groups was not largely different during the assays. When DA neuron differentiation was assessed at differentiation day 6 by the number of cells positive for tyrosine hydroxylase (TH), a key enzyme in DA biosynthesis, the number of TH+ cells was significantly greater in the cultures mixed with astrocytes compared with control cultures mixed with Ctx-NPCs (Figure 1, B and C). There was no significant difference in the TH+ cell yields promoted by coculturing with VM-Ast compared with those promoted by coculturing with Ctx-Ast (Figure 1C).
 Figure 1
Figure 1DA neuron differentiation and maturation promoted by coculturing with astrocytes. (A) Schematic for the coculture experiments. (B) Representative images of TH+ DA neurons differentiated from VM-NPCs in the presence of Ctx-NPCs (control), Ctx-Ast, or VM-Ast. Scale bar: 100 μm. Insets, enlarged images of the boxed areas (original magnification, ×400). (C) DA neuronal yields at differentiation day 6 (D6). (D–H) Morphometric measurement of DA neuron maturity assessed by neurite outgrowth lengths (D), soma size (E), and by the Sholl test (F and G). In the Sholl analysis, the total number of neurite crossings was counted in each circle with the radius increasing in steps of 15 μm (F). The critical value (G) is the radius at which there was a maximum number of neurite crossings. *P < 0.05, significantly different from the control; #P < 0.05, significantly different from Ctx-Ast, 1-way ANOVA. n = 3–5 wells (C) and 100 (D and E) and 25–30 (F and G) TH+ cells in each group.(H) Representative TH+ neuronal morphologies reconstructed in 3D by Neurolucida. (I–L) Synaptic densities on TH+ fibers. (I) Representative confocal images of synapsin+TH+ fibers. Scale bar: 25 μm. Puncta positive for the synaptic vesicle–specific markers SV2 (J), synapsin (K), and bassoon (L) on TH+ fibers were counted. *P < 0.05, significantly different from the control; #P < 0.05, significantly different from Ctx-Ast. n = 20 TH+ fibers for each group. (M) Presynaptic DA release. n = 3 independent cultures. DA levels were measured in the media conditioned in the differentiated cultures for 24 hours (D13–D15) and in cultures evoked by KCl-mediated depolarization for 30 minutes. *P < 0.05; #P < 0.05, 1-way ANOVA.
TH+ DA neurons differentiated in the presence of astrocytes exhibited more mature neuronal morphology than control cultures (Figure 1B, insets), which was quantified by neurite length (Figure 1D) and soma size (Figure 1E) of the TH+ cells. The morphometric maturity of TH+ DA neurons was further assessed based on the complexity of neurite outgrowth using the Sholl test (Figure 1, F and G). In contrast to the lack of difference in TH+ cell yields promoted by Ctx- versus VM-Ast, TH+ DA neurons cocultured with VM-Ast exhibited significantly greater neurite outgrowths than those cocultured with Ctx-Ast (Figure 1, B, D, F–H). In images captured by a confocal microscope, puncta positive for the synaptic vesicle–specific markers SV2, synapsin, and Bassoon were more abundantly localized in the neurites of the TH+ DA neurons differentiated with astrocytes, and the puncta densities were greatest in those cocultured with VM-Ast (Figure 1, I–L). The types of synapses formed were further analyzed by using neurotransmitter specific pre- and postsynaptic markers. Nigral mDA neurons in the midbrain physiologically receive glutamatergic excitatory and GABAergic inhibitory synaptic inputs from several regions of the brain (35). Excitatory postsynaptic density protein 95 (PSD95) clusters were found on dendritic shafts of TH+ DA neuronal cells, and many of them were juxtaposed with the vesicular glutamate transporter 2 (vGlut2) puncta, which indicates the presence of aspiny glutamatergic shaft synapses in TH+DA neurons (Supplemental Figure 2A). We found that vGlut2+PSD95+ clusters were more abundantly detected in TH+ DA neurons cocultured with astrocytes than in control cultures (Supplemental Figure 2, A and B). Along with significantly more vGlut2+PSD95+ clusters on TH+ cells cocultured with VM-Ast than those with Ctx-Ast (P = 00002), the glutamatergic synapses on spine-like structures were also found only with VM-Ast, further indicating more mature neuronal morphology with VM-Ast. GABAergic inhibitory (vGAT+Gephyrin+) synaptic puncta were also more abundantly localized in the fibers of TH+ DA neurons cultured with astrocytes (Supplemental Figure 2, A and B). We further observed that DA release, especially when evoked by KCl-induced depolarization, was significantly promoted in the cultures differentiated with astrocytes; the presynaptic DA release promoted by astrocytes was greater in the cultures with VM-Ast than in the cultures with Ctx-Ast (Figure 1M).
mDA neurons are specified based on the expression of several midbrain-specific genes, such as Nurr1, Foxa2, Lmx1a/b, Pitx3, and En1/2. As these midbrain markers play critical roles in DA phenotype maintenance, survival, and function (36–39), their expression is critical in the preparation of mDA neurons for therapeutic purposes. However, we and others have recently shown that the expression of midbrain-specific markers in mDA neurons is largely affected by in vitro and in vivo environments and thus easily lost during passages, long after differentiation in vitro and after transplantation in vivo (16, 40, 41). Indeed, expression of Foxa2, Nurr1, and Lmx1a was detected only in 64%, 44%, and 46% of TH+ cells at day 12 of differentiation, respectively (Figure 2, A and B). The percentages of TH+ DA neurons expressing the midbrain markers (Figure 2B) and their expression levels in individual TH+ cells (estimated by mean fluorescence intensity [MFI]) in the nucleus of TH+ cells (Figure 2C) were significantly greater in the cultures mixed with astrocytes. The coculture with VM-Ast had the greatest effect, with almost all TH+ cells coexpressing the midbrain-specific markers, further supporting the functional superiority of VM-Ast over Ctx-Ast in their midbrain dopaminotrophic actions.
 Figure 2
Figure 2DA neurons differentiated with astrocytes are more resistant to toxic insult along with increased midbrain-specific factor expressions. (A–C) Expression of midbrain-specific markers in the differentiated DA neurons. Expression levels of the midbrain-specific markers in C were determined in individual TH+ DA neurons by measuring MFI using LAS image analysis (Leica). *P < 0.01; #P < 0.01, 1-way ANOVA. n = 9 microscopic fields (B) and n = 32–36 TH+ cells (C). Scale bar: 50 μm. (D–F) Resistance of DA neurons against a toxic stimulus. TH+ DA neurons differentiated in the presence of Ctx- or VM-Ast (Ctx-NPCs as the control) at D12 were exposed to H2O2 (1,000 μM) for 10 hours and viable TH+ cell counts (E) and fiber lengths (F) were measured on the following day. Representative TH+ cell images after H2O2 treatment are shown in D. Scale bar: 100 μm. Insets, enlarged images of the boxed areas (original magnification, ×400). Data in E represent percentage of TH+ cells that survived after the toxin exposure compared with those that were not exposed to toxin (H2O2 = 0 μM). *P < 0.01; #P < 0.01, 1-way ANOVA. n = 3 independent experiments (2–3 culture wells/experiment) (E) and n = 80–90 TH+ cells (F).
When the differentiated cultures were exposed to the ROS-producing agent H2O2 (1000 μM for 10 hours), only 15% of the toxin-untreated TH+ DA neurons survived in the control cultures mixed with Ctx-NPCs (Figure 2, D and E). As expected, remarkably lower proportions of DA neurons died after the toxic stimulus in the presence of astrocytes. In addition, TH+ cells in the cultures mixed with astrocytes survived the toxin treatment and displayed healthy neuronal morphology with extensive neurite outgrowths, while most of the surviving TH+ cells in the control cultures had blunted or fragmented neurites (Figure 2D, insets), a neuronal aging and degenerative phenotype (42). This finding was further supported by quantification of the fiber length in the surviving TH+ cells (Figure 2F). Again, VM-Ast exerted significantly greater protective effects on mDA neurons than Ctx–astrocytes (Figure 2, D–F). Collectively, these findings suggest that cultured astrocytes facilitate VM-NPC differentiation toward authentic mDA neurons that are morphologically, synaptically, and functionally mature and are resistant to toxic insults. In addition, our data show that astrocytes of VM origin are superior to astrocytes from the Ctx in terms of their trophic actions on mDA neuron differentiation and survival.
Paracrine manner of the observed astrocytic actions. The observed astrocytic actions could be mediated by factors released from the astrocytes and/or cell-cell contact signaling. To test the possibility of paracrine effects, the medium was conditioned in the astrocyte cultures (or differentiated Ctx-NPC cultures as the control) for 2 days and the conditioned medium (CM) was added to undifferentiated VM-NPC cultures (Figure 3A). The astrocyte-CM more efficiently promoted VM-NPC differentiation into DA neurons compared with the control-CM,as estimated by TH+ cell yields 6 days after differentiation (Figure 3, B and C). In addition, fiber outgrowth (Figure 3D) and branching (Figure 3E) during the 42 hours of CM treatment increased in the differentiating cultures treated with astrocyte-CM compared with the control-CM, with the greatest increase by VM-Ast-CM treatment. Along with the morphologic maturation effects, similar patterns of the astrocyte-CM effects were manifested in the expression of NeuN, a mature neuron marker, in TH+ DA neuronal cells (Figure 3, F, I, and J). Expression of the midbrain-specific factors Nurr1 and Foxa2 (Figure 3, G–J) and TH+ cell survival with H2O2 toxin treatment (Figure 3, K–M) were greatly improved by VM-Ast-CM treatment, compared with that in cultures treated with control-CM or Ctx-Ast-CM. These findings collectively indicate that the astrocyte functions observed in Figures 1 and 2 were at least in part mediated by paracrine factors secreted from the astrocytes.
 Figure 3
Figure 3Paracrine manner of the astrocyte-mediated neurotrophic actions. (A) Schematic of the CM treatment experiments. Media conditioned in Ctx- or VM-Ast (or Ctx-NPC as control) was collected and added to cultured VM-NPCs during the differentiation period. (B) Representative images for TH+ DA neurons at D12. Scale bar: 100 μm. Insets, enlarged images of the boxed areas (original magnification, ×400). (C) TH+DA neuronal yields at D6. *P < 0.05, significantly different from control. n = 3 cultures for each group. (D and E) Morphometric measurement of neurite outgrowth assessed by a time-lapse imaging system. VM-NPCs at D3 were treated with the CMs. Neurite lengths (D) and branch points of the neurites (E) in 27 randomly selected microscopic fields from 3 independent cultures were automatically analyzed for 42 hours using IncuCyte’s NeuroTrack software. (F–J) Expression of mature neuronal (NeuN) and mDA neuronal (Foxa2, Nurr1) markers in the differentiated DA neurons. I and J depict percentage of TH+ cells expressing the markers and the expression levels (MFI) in individual TH+ DA neurons, respectively. *P < 0.01, significantly different from control; #P < 0.01, significantly different from Ctx-Ast, 1-way ANOVA. n = 9 microscopic fields (I) and n = 32–36 TH+ cells (J). (K–M) Resistance of DA neurons against a toxic stimulus. TH+ DA neurons differentiated in the presence of Ctx- or VM-Ast-CM (Ctx-NPC-CM as the control) at D12 were exposed to H2O2 (1,000 μM) for 8 hours and viable TH+ cells were counted on the following day (L). Shown in K are representative TH+ cell images after H2O2 treatment. Scale bars: 100 μm. Insets, high-powered images of the boxed areas (original magnification, ×400). Fiber lengths of surviving TH+ cells were also estimated (M). *P < 0.01; #P < 0.01, 1-way ANOVA. n = 3 independent experiments (2–3 wells/experiment) (L) and n = 80–90 TH+ cells (M). (N) Expression of neurotrophic genes in the cultured Ctx-Ast, VM-Ast, and Ctx-NPCs, estimated by real-time PCR (qPCR) analyses. *P < 0.01; #P < 0.05, 1-way ANOVA. n = 3.
We next sought to identify the paracrine factors responsible for the astrocyte-mediated neurotrophic actions. Consistent with neurotrophic factor secretion from astroglial cells (43), cultured astrocytes expressed elevated levels of mRNAs for various neurotrophic factors (Figure 3N). Of note, the expression of glial cell–derived neurotrophic factor (Gdnf), sonic hedgehog (Shh), and Fgf8 was greatly elevated in VM-Ast compared with Ctx-Ast or control Ctx-NPCs. GDNF has physiologic neurotrophic roles specific for mDA neurons (44), and SHH, by establishing an autoregulatory loop with Foxa2, is one of the most important factors in mDA neuron development and survival (45, 46). The cooperative actions of FGF8 and SHH are critical for mDA neuron development (47), and thus, this cytokine combination is commonly used for in vitro mDA neuron patterning from stem cells (48, 49). In addition, crucial roles for Wnt/β-catenin signaling from mDA neurogenesis to regeneration and secretion of Wnt1 and Wnt5a proteins from VM-Ast have been reported (50, 51). Consistently, cultured VM-Ast abundantly expressed not only Wnt cytokines (Wnt1, Wnt4, Wnt5a), but also spondin-2 (SPO-2) (Figure 3N), a secreted protein of the R-spondin family, which activates Wnt/β-catenin signaling by preventing clearance of the Frizzled–LRP Wnt receptor complex (52). In addition, cultured astrocytes exhibited an elevated expression of thrombospondin-1 (Thbs-1), Glypicans (GPC) 4 and 6, and secretory ECM proteins, which promote synapse formation (53, 54); Thbs-1 mRNA levels were higher in VM-Ast cultures than in Ctx-Ast, suggesting that these factors were responsible for the synaptic maturation of DA neurons in the presence of VM-Ast (Figure 1, I–L, and Supplemental Figure 2).
Differentially expressed genes among the differentiated NPCs, Ctx-Ast, and VM-Ast. To gain further molecular insight into the observed astrocyte functions, we performed RNA-seq analysis of the differentiated Ctx-NPCs (control), Ctx-Ast, and VM-Ast used in the coculture and CM experiments. The differentially expressed genes (DEGs) in Ctx-Ast compared with differentiated Ctx-NPCs (FPKM > 1, log2 > 1) significantly overlapped with DEGs in VM-Ast compared with differentiated Ctx-NPCs (N-1′ χ2 = 9059.4, df = 1, P < 2.2 × 10–16, Figure 4A). In a gene set analysis, the overlapped genes (hereafter, referred to as common astrocytic genes) were enriched in the gene ontologies associated with cell adhesion/ECM, immune/inflammatory response, and neuron differentiation (Figure 4B). Surprisingly, the top-ranked gene ontologies for DEGs between Ctx-Ast and VM-Ast also fell within similar gene categories (Figure 4, C and D). The gene sets enriched in cultured astrocytes (versus control NPCs) and VM-Ast (versus Ctx-Ast) were further confirmed in gene-set enrichment analysis (GSEA) (Supplemental Figure 3A). Specifically, the differential expression patterns for the neurotrophic factors determined by quantitative PCR (qPCR) (Figure 3N) were similarly replicated in the RNA-seq analyses (Figure 4E), indicating fidelity of the RNA-seq analysis and further confirming that these neurotrophic factors are responsible for the observed trophic effects of astrocytes. Cell adhesion/ECM that was annotated to one of the top-ranked ontologies is of prime interest due to the importance of cell-to-cell and cell-to-ECM contacts in stem cell behaviors and regenerative processes in damaged tissues (55–57). The heatmap for the cell adhesion/ECM molecules exhibiting increased levels of expression in VM-Ast (compared with Ctx-Ast and/or control NPCs) is shown in Figure 4E and Supplemental Figure 3B. Of note, among these genes, the upregulated expression of fibronectin 1 (FN1) and integrin (Itgb4) in astrocytes (VM-Ast) seems to be based on a recent study reporting that successful engraftment of DA neurons requires cell-to-ECM adhesion through the interaction between integrin and fibronectin in an animal model of PD (58). In addition, tenascin (Tnc), another upregulated gene in VM-Ast, has been reported to augment grafted DA neuron attachment and survival after brain injury and transplantation (59). CSPGs are known to inhibit axonal regeneration and neurogenesis (60). Notably, the expression of Neurocan (Cspg3) and Brevican (Cspg7) was greatly downregulated in the VM-Ast cultures compared with Ctx-Ast and control NPC cultures (Figure 4E). In addition, the cultured Ctx- and VM-Ast commonly showed a remarkable reduction in the expression of myelin-basic protein (Mbp) and myelin-associated protein (Mag), which inhibits neurite outgrowth (61), and myelin oligodendrocyte glycoprotein (Mog35-55), a peptide causing neuroinflammation (62).
 Figure 4
Figure 4Potential molecules mediating the dopaminotrophic functions of astrocytes. (A–E) mRNA-seq analyses for the DEGs among VM-Ast, Ctx-Ast, and control Ctx-NPCs. Genes upregulated and/or downregulated by over 2-fold were selected for analysis. (A) Venn diagram summarizing the overlap between DEGs from VM-Ast versus Ctx-NPCs (left circle) and Ctx-Ast versus Ctx-NPCs (right circle). The numbers of genes are indicated in the Venn diagram. To calculate overlap with DEGs, statistical analysis was performed using the χ2 test based on a 2 × 2 table using R version 3.3.2 of the MASS package. (B) GO and KEGG analyses for the 4,445 overlapping genes (common astrocytic genes). Purple bars indicate the number of genes under the designated GO term/KEGG pathway. Yellow bars indicate P values, and negative logs of the P values (bottom) are plotted on the x axis. (C and D) Volcano plot (C) and GO/KEGG analyses (D) for the DEGs between VM-Ast and Ctx-Ast. (E) Heatmap showing fold changes of the selected genes in VM- and Ctx-Ast compared with control Ctx-NPCs (log2 folds, VM-Ast/Ctx-NPC and Ctx-Ast/Ctx-NPC). (F) Expression of important pro- and antiinflammatory and secretory antioxidant genes was further confirmed by qPCR analyses. *P < 0.05, significantly different from control; #P < 0.05, significantly different from Ctx-Ast, 1-way ANOVA. n = 3 PCR reactions.
The enrichment of DEGs in the category of immune/inflammatory response in astrocytes compared with control NPCs is not surprising, considering that astrocytes are commonly believed to be a cell type mediating brain inflammation. Expression of proinflammatory cytokine mRNAs was elevated in astrocyte cultures, and interestingly, levels were higher in VM-Ast than in Ctx-Ast (Figure 4E and Supplemental Figure 3, A–C). Proinflammatory cytokines can trigger cellular defense mechanisms and are frequently linked to enhanced neuronal differentiation and survival (63–66). Despite these positive aspects, the common idea is that proinflammatory cytokines establish a cytotoxic inflammatory milieu. Along with the proinflammatory gene-expression increase, gene expression of antiinflammatory and neurotrophic glial markers also increased in the astrocyte cultures (Figure 4E and Supplemental Figure 3B). For example, the gene with the top-ranked expression enriched in the VM-Ast cultures was Il-19, an antiinflammatory cytokine from the IL-10 family (Supplemental Figure 3F). The mRNA expression levels of the other antiinflammatory markers arginase 1 (Arg1, an enzyme inhibiting NO biosynthesis) and IL-1 receptor antagonist (IL1RN) were much higher in the cultured VM-Ast than in Ctx-Ast and control NPC cultures: FPKMs of ARG1 were 49.9, 5.5, and 0.03, and FPKMs of IL1RN were 14.0, 1.4, and 0.04 for VM-Ast, Ctx-Ast, and control NPCs, respectively (Figure 4E). The expression of the antiinflammatory cytokines Il-10 and Il-27 was the highest and only detected in the VM-Ast cultures, although they were excluded from the gene set analysis due to low FPKM values (<1). IFN-α and -β exert antiinflammatory (67, 68) and neuroprotective activities against mDA neuronal degeneration (69). It has recently been reported that antiinflammatory actions of type I IFNs are mediated via the aryl hydrocarbon receptor (Ahr) in astrocytes (67). Of note, along with the increase in type I IFN expression in qPCR analyses (Figure 4F), the AHR expression level in VM-Ast cultures was also greatly unregulated (FPKM: 51.34 [VM-Ast], 6.21 [Ctx-Ast], and 3.21 [differentiated (diff.) NPC ]) (Figure 4E), indicating that the expression of these antiinflammatory cytokines/receptors contributes to improved cell survival by mitigating proinflammatory cytotoxic effects in the cultures cocultured with VM-Ast. The increase in expression levels of pro- and antiinflammatory genes was further confirmed by qPCR analyses (Figure 4F). When gene enrichment analysis was performed on the genes, expressions of which were lower in VM-Ast (vs. Ctx-Ast), the category ion channels was the top-ranked ontology (Supplemental Figure 3, D and E). An expanding idea is that ion channels in astrocytes contribute to astrocyte biology and crosstalk between neurons and glia (70). Thus, the differential expression of ion channels is likely to be valuable in the understanding of their region-specific functions.
The expression of multiple antioxidant genes was upregulated in the cultured astrocytes compared with control NPCs (Figure 4E). However, ROS-scavenging activity, as estimated by intracellular glutathione levels, was relatively lower in the cultured astrocytes than in the control cultures (Supplemental Figure 4A). Another neuroprotective mechanism of astrocytes occurs via clearance of glutamate-induced toxicity (71). Along with enriched expression of the glutamate transporters GLAST and GLT-1 in cultured astrocytes (Supplemental Figure 1, B and C), glutamate uptake activities were greater in cultured astrocytes than in the control NPCs (Supplemental Figure 4B). Notably, the much greater glutamate uptake in cultured VM-Ast compared with Ctx-Ast suggests that the enriched glutamate clearance activity contributes to the observed neuroprotective actions mediated by astrocytes, especially VM-Ast.
Forced Nurr1+Foxa2 expression further potentiates the neuroprotective actions of VM-Ast. Next, we investigated whether forced Nurr1+Foxa2 expression in VM-Ast further promotes the astrocyte-mediated dopaminotrophic actions. VM-Ast were transduced with Nurr1+Foxa2-expressing lentiviruses or mock viruses (control). The VM-NPCs harvested were cocultured with the transduced astrocytes, and the NPC behaviors associated with their therapeutic capacity upon transplantation were assessed (Figure 5A). DA neuronal yield (Figure 5B), morphologic (TH+ fiber lengths, Figure 5C) and synaptic maturation (SV2+ puncta density, Figure 5D), and the expression of midbrain-specific markers (Figure 5E) in the DA neurons differentiated from VM-NPCs in the presence of Nurr1+Foxa2-transduced VM-Ast (N+F-Ast) were indistinguishable from those in cultures cocultured with the control-Ast. However, presynaptic DA neuronal functionality, as estimated by DA neurotransmitter release, was significantly greater in the cultures differentiated in the presence of N+F-Ast than in cultures differentiated with the control-Ast (Figure 5F). In addition, differentiated TH+ DA neurons in the presence of N+F-Ast were more resistant to the toxic insult induced by H2O2 treatment than differentiated TH+ DA neurons cocultured with the control-Ast (Figure 5, G and H). A larger number of TH+ DA neurons along with healthier neuronal shape were also observed in the cultures treated with the medium conditioned in N+F-Ast (N+F-CM) compared with cultures treated with control-CM (Figure 5, I–L), suggesting that the Nurr1+Foxa2 neuroprotective actions were mediated in a paracrine manner.
 Figure 5
Figure 5Forced expression of Nurr1+Foxa2 in VM-Ast potentiates astrocyte-mediated dopaminotrophic actions. (A) Schematic of procedure of the coculture experiments. VM-Ast were transduced with Nurr1+Foxa2-expressing lentiviruses (or control mock viruses), harvested, and mixed with VM-NPCs. After differentiation was induced in the VM-NPCs mixed with the astrocytes, TH+ DA neuronal yields (B), morphologic (C) and synaptic maturation (D), expression of midbrain-specific markers (E), presynaptic DA release (F), and resistance to a toxic stimulus (G and H) were assessed in the TH+ DA neurons in the differentiated cultures. (I–L) CM treatment experiments. CM was prepared from Nurr1+Foxa2-transduced VM-Ast (or mock-transduced VM-Ast as control) and added to VM-NPCs during the differentiation period. The differentiated cultures at D12 were exposed to H2O2 (1,000 μM, 10 hours), and the number (K) and neurite length (L) of surviving TH+ cells were estimated as described in the Figure 3 legend. *P < 0.05; #P < 0.05, 2-tailed Student’s t test. n = 3 culture wells (F, H, K) and 80–90 TH+ cells (L). Scale bars: 100 μm.
In RNA-seq analysis of the N+F-Ast compared with control-Ast, the top 10 ranked ontologies enriched by the DEGs included immune/inflammation, response of wound healing, and cell adhesion (Figure 6A). In addition, when we performed gene set analysis with genes with downregulated expression in N+F-Ast versus control-Ast (log2 < –1), 4 out of the top 5 ontologies were related to immune/inflammation (Figure 6, B and D), which was further confirmed by GSEA with curated gene sets immune response and inflammatory response (Figure 6C), indicating that Nurr1+Foxa2 expression in the VM-Ast may mainly exert extrinsic neuroprotective roles by reducing immune/inflammation-mediated neurotoxicity. Specifically, decreased mRNA levels of proinflammatory cytokines (Il-1b, Il-6, Tnfa, and iNOS) and myelin-associated proteins (Mbp, Mag, and Mog) were manifested in the RNA-seq data (Figure 6E) and those were further confirmed by qPCR analyses (Figure 6F). In addition, increased expression of secretory neurotrophic (Shh, Bdnf, Gdnf, and Nt3) and antiinflammatory (Ifna, Ifnb) factors was also detected in N+F-Ast (Figure 6F), suggesting that they also contributed to the N+F-CM–mediated neuroprotective actions.
 Figure 6
Figure 6RNA-seq analysis for the DEGs between Nurr1+Foxa2- and control VM-Ast. (A) GO and KEGG analyses for the genes upregulated (log2 > 1) or downregulated (log2 < –1) in Nurr1+Foxa2-VM-Ast (NF-Ast) compared with control VM-Ast (Cont-Ast) (FPKM > 1). (B) Gene ontologies only for the genes downregulated in NF-Ast vs. control-Ast (log2 < –1, FPKM>1). (C) GSEA for immune response and inflammatory response gene sets enriched in Nurr1+Foxa2-VM-Ast versus VM-Ast. (D) Heatmap of the expression levels (FPKMs) of all the genes annotated to the ontologies related to immune (121 genes)/inflammatory response (103 genes) in control-Ast vs. NF-Ast in B. Expressions of selected immune/inflammatory, inhibitory ECMs, and antioxidant genes are further shown in E. (F) mRNA levels of important pro- and antiinflammatory and neurotrophic factors were further confirmed by real-time qPCR analyses. *P < 0.05, 2-tailed Student’s t test. n = 3.
Further examination of the RNA-seq data identified multiple antioxidant enzymes with upregulated gene expression in N+F-Ast (Figure 6E). Consistently, significantly greater intracellular glutathione levels were manifested in the N+F-VM-Ast compared with the control VM-Ast cultures (Supplemental Figure 5A). Consistent with the upregulated secretory ROS scavenging factors Sod3 and Gpx3 (Figure 6E), ROS levels in mDA neuron–enriched cultures (differentiated from VM-NPCs) were dramatically decreased by N+F-CM treatment (Supplemental Figure 5B). These findings collectively indicated that the enhanced ROS scavenging capacity in VM-Ast by Nurr1+Foxa2 also contributed to N+F-Ast–mediated neuroprotective actions. Glutamate uptake activity, however, was indistinguishable between control and N+F-VM-Ast (Supplemental Figure 5C).
Cografting of astrocytes potentiates the cell therapeutic effects of VM-NPC transplantation via establishing a neurotrophic host brain environment. Although our in vitro data clearly supported the neurotrophic actions of cultured astrocytes, it remains to be determined whether similar astrocyte-mediated effects occur after transplantation in the host brain in vivo, where the grafted cells are inevitably exposed to hostile inflammatory/immunogenic environments and interact with endogenous cells such as glia, neuronal cells, and peripheral blood cells that may enter through the disrupted blood-brain barrier (BBB) during the cell-injection process. Of note, it has been reported that glia can be reactivated into the detrimental phenotype during a delayed injury phase (72–74), raising concerns that grafted astroglia may also convert into the harmful phenotype after transplantation. Thus, our first round of transplantation experiments was performed to determine whether transplantation of cultured astrocytes could establish a neurotrophic environment. Similarly to what was shown in the in vitro data, the expression of neurotrophic factors (Gdnf, Nt3, Shh, Wnt1, Wnt3, Wnt5), trophic ECM proteins (Col6a2, Fn1, Thbs-1), and antioxidant proteins (Gpx3 and Sod3) was upregulated in the striatum transplanted with VM-Ast compared with the striatum grafted with control Ctx-NPCs (or Ctx-Ast) at 1 month after transplantation (Figure 7A). In addition, the expression of antiinflammatory phenotype markers (Ifnb, Ccl17, Il-1r2, Il-1rn, Ym1, and Il-10) was also greater in brains grafted with VM-Ast. Notably, in contrast with what was seen in the in vitro findings, the expression of proinflammatory phenotype genes did not increase, and some genes (iNOS, Il-1β, Cxcl11) were downregulated in brains grafted with VM-Ast compared with brains grafted with control NPCs (Figure 7A), suggesting that the naive VM-Ast property of high expression of proinflammatory genes in vitro was altered in the grafted in vivo brain environments, probably due to the interaction with endogenous cells. It is also possible that endogenous microglia/astrocytes were activated into the antiinflammatory/neurotrophic phenotype due to their interaction with grafted VM-Ast. We further confirmed the astrocyte-grafting effects by immunohistochemical analyses (Figure 7B). Cells (astrocytes, microglia) immunoreactive for the markers specific for antiinflammatory/neurotrophic glia (Arg1, CD206) were significantly greater (P < 0.00066) in the graft-host interfaces of the brain transplanted with cultured astrocytes, compared with those grafted with Ctx-NPCs. Reduction of the cells expressing proinflammatory/cytotoxic factors (iNOS, CD11b, CD16) also occurred. An upregulated expression pattern of neurotrophic, antioxidant, and ECM genes was further observed in the brains grafted with N+F-transduced VM-Ast compared with brains transplanted with control VM-Ast (Figure 8A). The phenotype transition by N+F-VM-Ast transplantation was further clarified in immunohistochemical analyses exhibiting downregulated expression of proinflammatory/cytotoxic phenotype markers and upregulated expression of antiinflammatory/neurotrophic phenotype markers in the brains transplanted with N+F-VM-Ast compared with brains grafted with control VM-Ast (Figure 8B). Taken together, these findings indicate that transplantation of cultured VM-Ast could establish a neurotrophic brain environment and that Nurr1+Foxa2 expression in the donor astrocytes potentiates the astrocyte transplantation effect.
 Figure 7
Figure 7Improvement of host brain environments by grafting cultured astrocytes. Rats were transplanted with VM-Ast, Ctx-Ast, or Ctx-NPCs (control). (A) qPCR data for gene expressions associated with neurotrophic or hostile brain environments among the grafted groups. The PCR analyses were carried out in the graft-host interfaces dissected as described in Methods at 1 month after transplantation. *P < 0.05; #P < 0.05. n = 3. (B) Immunohistochemical analyses for the glial cells immunoreactive for proinflammatory/cytotoxic (iNOS, CD11b, CD16) and antiinflammatory/neuroprotective (Arginase, CD206) factors. The immunoreactive cells along the graft-host interfaces were counted in 6 cryosectioned slices from 3 animals/each group at 7–10 days after transplantation. Data are expressed as percentage of immunoreactive cells out of DAPI+ cells, of GFAP+ astrocytic, and of Iba1+ microglial populations. *P < 0.05; #P < 0.05, 1-way ANOVA. n = 6. Scale bar: 100 μm.
 Figure 8
Figure 8Effects of Nurr1+Foxa2 priming in donor VM-Ast to improve host brain environments after transplantation. Neurotrophic or inflammatory factor expressions in the brains grafted were determined by qPCR (A) and immunohistochemical (B) analyses as described in Figure 7. *P < 0.05; #P < 0.05, 2-tailed t test. n = 3 PCRs (A) and 6 slices (B). Scale bar: 100 μm.
We ultimately assessed whether cografting astrocytes could improve cell therapeutic outcomes in PD. VM-NPCs mixed with cultured VM- or Ctx-Ast (or Ctx-NPCs as control) were intrastriatally transplanted into a hemi-parkinsonian rat model, and PD-related behaviors were assessed for 6 months after transplantation. An amphetamine-induced rotation test revealed only a minor reduction in rotation scores compared with pretransplantation values in PD rats cografted with VM-NPCs+Ctx-NPCs (control), but greater behavioral recovery was achieved by cografting with astrocytes (Figure 9A). Especially, cografting of N+F-VM-Ast along with VM-NPCs resulted in an almost complete recovery in the rotation scores (>95% reduction in rotation score from 3 months after transplantation, n = 6). We also assessed PD behaviors using nonpharmacological assays at 6 months after transplantation. Similar patterns of behavioral recovery among the animal groups were observed in the step adjustment (Figure 9B) and cylinder tests (Figure 9C). Consistent with the data from the in vitro coculture and CM experiments as well as the behavioral assays, histologic analyses performed 6 months after transplantation exhibited formation of a much larger TH+ cell graft in rats cotransplanted with astrocytes than with control NPCs (Figure 9, D and E). All indices for DA neuron engraftment (Figure 9, E–G) were significantly greater in animals cografted with VM-Ast compared with Ctx-Ast, but no significant difference in the indices was detected between the animal groups cografted with VM-Ast and those cografted with N+F-VM-Ast. TH+ cells in the striatum cografted with astrocytes exhibited healthier and more mature neuronal maturity than those in the control grafts (Supplemental Video 1 and Figure 9, H and I), which was confirmed by the estimation of TH+ fiber length (Figure 9J) and synapsin+ puncta density (Figure 9, K and L). Only a slight but significant increase in the TH+ fiber lengths was detected in the N+F-VM-Ast–cografted groups compared with those cografted with the control-VM-Ast (Figure 9J). Surprisingly, the midbrain-specific markers Nurr1 and Foxa2, expression of which is easily lost in donor cells after transplantation (16, 40, 41), were faithfully colocalized in TH+ DA neurons in the grafts generated by astrocyte cotransplantation, especially by cografting with N+F-VM-Ast, even 6 months after transplantation (Supplemental Figure 6, A–C). TH+ DA neuronal cells in the grafts were surrounded by GFAP+ astrocytes (probably both transplanted and endogenous) (Supplemental Figure 6D) and Iba1+ microglia (endogenous) (Supplemental Figure 6E). The Iba1+ microglia were ramified with a resting or neuroprotective morphology (75), indicating that the neighboring and covering of the grafted DA neurons were protective. These findings collectively indicated that cotransplantation of VM-NPCs with astrocytes, especially with N+F-VM-Ast, ensured a long-term engraftment of mature authentic mDA neurons via astrocytic actions to improve host brain environments (schematized summary is shown in Figure 10).
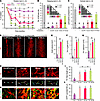 Figure 9
Figure 9Cografting of astrocytes improves the therapeutic effects of VM-NPC transplantation in a PD rat model. NPCs derived from rat embryonic VM at E12 were expanded in vitro, harvested, and mixed with Ctx-NPCs (control), Ctx-Ast, VM-Ast, or N+F-VM-Ast before cell injection. The mixed cells were intrastriatally transplanted into 6-OHDA–lesioned PD model rats. Behavioral (A–C) and histological (D–L) analyses were carried out every month for the 6 months after transplantation (A) or at 6 months after transplantation (B–L). (A) Amphetamine-induced rotation scores were determined every month for the 6 months after transplantation. Data are given as percentage changes in rotation scores for each animal compared with the pretransplantation value. Mean + SEM of the rotation scores is depicted. n = 6 for each group. Significant decreases in rotation scores were seen in animals that received astrocyte cografts compared with those that received control NPCs alone. *P < 0.05; #P < 0.05, Ctx-Ast; †P < 0.05, VM-Ast at each posttransplantation time point, 1-way ANOVA. Behaviors of the transplanted animals were further assessed by step adjustment (B) and cylinder (C) tests at 6 months after transplantation. Statistical significances (P < 0.01) among groups are expressed using schematics in the graphs. One-way ANOVA followed by Bonferroni’s post hoc analysis. (D–L) Histologic analyses 6 months after transplantation. (D) Overview of the TH+ cell grafts. (E) Graft volume. (F) Total number of TH+ cells. (G) TH+ cell density in the graft. *P < 0.05; #P < 0.05, 1-way ANOVA. (H–J) Morphologic maturation of DA neurons in the grafts estimated by TH+ fiber length. Shown are immunohistochemical (H) and Neurolucida reconstruction of representative TH+ neuronal images (I). (K and L) Synaptic maturation of TH+ DA neurons estimated by synapsin+ puncta density. *P < 0.05; #P < 0.05, 1-way ANOVA. Scale bars, 100 μm (D); 50 μm (H); 25 μm (K).
 Figure 10
Figure 10Schematic summary for the trophic actions of astrocytes cografted in PD cell therapeutic approach. Astrocytes (VM, N+F-VM-Ast) cografted with VM-NPCs promote a series of transplanted NPC survival, mDA neuron differentiation, neuronal maturation, and synaptic integration by correcting hostile host brain environments. The astrocytic actions are attained via secretion of various neurotrophic ECM proteins, antioxidants, neurotrophic cytokines, GLAST/GLT1-mediated clearance of glutamate toxicity, etc. Ultimately, PD behaviors are improved along with enriched engraftment of mature and functional mDA neurons expressing midbrain-specific markers.
-
Discussion
It has long been suggested that after transplantation, the host brain becomes hostile to grafted cells and that this detrimental host brain environment is mainly responsible for unsatisfactory cell transplantation therapeutic results with poor neuronal engraftment. Nevertheless, targeting the host brain to improve cell therapeutic effects has only been addressed in a few studies (58, 76). Furthermore, none of these studies used an approach to fundamentally correct inflammatory cytotoxic host brain environments. Based on the physiologic neurotrophic functions of astrocytes, utilization of this cell type is a potential strategy to modify pathologic brain environments. However, few studies have examined developing an astrocyte-based therapy for neurologic disorders, mainly because of the potential reactivation of this cell type into a detrimental/neurotoxic phenotype in diseased brains (13). However, cumulative studies have demonstrated that cultured astrocytes generally exhibit immature properties, as shown in Supplemental Figure 1, D–H, of our study, and that transplanted immature astrocytes do not undergo harmful reactivation after CNS injury, but rather support neurite outgrowth and reduce glial scar formation in the injured CNS (reviewed in ref. 77). Based on these findings, we attempted cografting with astrocytes to attain an improvement in therapeutic efficacy of cell transplantation for PD.
As shown in Supplemental Figure 1A, astrocyte cultures are frequently contaminated with microglia. Microglia induce proinflammatory/cytotoxic reactivation of astrocytes (13). In addition, it has been reported that neurotrophic functions of astrocytes also require minor populations of microglia (<1%) (78). To determine whether microglia contamination affects neurotrophic functions of astrocytes, we generated microglia-free astrocyte cultures by modifying the culture protocol (including one round of mild trypsin treatment step; see Methods). mDA neuronal differentiation, morphologic maturation, and toxin resistance promoted by CM derived from pure astrocyte cultures were comparable (or slightly lower) to those promoted by the CM from the astrocyte cultures containing the minor microglia population (Supplemental Figure 7), indicating that astrocytes in culture can exert the observed trophic actions in the absence of microglia and that minor microglia contamination (in this study, <0.5%) also does not affect the naive neurotrophic actions of astrocytes. Whether further, higher levels of microglia contamination are detrimental or beneficial remains to be determined.
Like neurons, astrocytes are thought to have regional identities and play region-specific roles. Thus, astrocytes cultured from the VM neurogenic niche were expected to be dopaminotrophic. Previous studies have consistently demonstrated that astrocytes derived from the VM facilitate DA neuron differentiation and survival via secretion of GDNF, FGF2, and Wnts (reviewed in ref. 66). In this study, we carried out further systematic and comparative analyses both in vitro and in vivo after transplantation to test the dopaminotrophic functions of cultured VM-Ast in comparison with astrocytes cultured from the nondopaminergic region of the Ctx and the effects of priming the VM-Ast with Nurr1+Foxa2, which has been reported to potentiate the neurotrophic functions of glia (16). In the niche established by the VM-Ast, grafted NPCs efficiently differentiated into morphologically, synaptically, and functionally mature mDA neurons and the differentiated mDA neuronal cells survived for long periods after transplantation while maintaining expression of midbrain-specific markers. The expression of midbrain-specific factors such as Nurr1 and Foxa2 is a critical indicator for successful mDA neuron engraftment (49), as the expression of these genes is required for mDA neuron survival, function, and phenotype maintenance (36–39). Considering that the expression of midbrain factors is easily lost in stressful conditions (16, 40, 41), sustained expression of midbrain markers in the presence of astrocytes is likely to be attained by the observed astrocyte actions that change the hostile inflammatory host brain milieu into a neurotrophic environment. Mechanisms underlying the astrocyte-mediated dopaminotrophic niche included secretion of the reported neurotrophic factors as well as other cytokines, such as SHH and FGF8, which had much greater levels of expression in cultured VM-Ast than in Ctx-Ast. Based on the previous findings (45, 46), it is likely that the expression of SHH and FGF8 is regulated by the midbrain-specific transcription factors Foxa2 and Lmx1a, respectively, expressions of which were maintained at high levels in cultured VM-Ast in a positive regulatory loop in VM-Ast. In addition, VM-Ast exhibited increased synthesis of various cell-cell contact and ECM molecules, including synaptogenic ECMs thrombospondins and glypicans, and enhanced glutamate clearance activity, both of which were superior to those in Ctx-Ast. Engineering of the midbrain-specific factors Nurr1+Foxa2 in the VM-Ast further improved their neuroprotective action mainly by reducing inflammation as well as by enhancing ROS-scavenging activity.
Based on the observed Nurr1+Foxa2 functions, priming these factor expressions in astrocytes is highly suggested prior to cografting. However, currently available methods for exogene expression, including lentiviral transduction used in this study, are toxic to cells and thus may reduce naive cellular functions. Indeed, a side-by-side comparative analysis has shown that DA release and resistance of DA neurons against H2O2 toxin were significantly lower in the mDA neuron cultures treated with the CM prepared in mock-transduced VM-Ast than in those treated with CM from nontransduced VM-Ast along with decreased neurotrophic factor expressions in the viral transduced astrocytes (Supplemental Figure 8). Forced Nurr1+Foxa2 expression effects were dramatic and sufficiently overcame the viral transduction–mediated reduction of astrocyte functions and also showed greater neurotrophic actions than the effects of the nontransduced control in all the assays. Nevertheless, development of an exogene expression system with minimal side effects is required to maximize the Nurr1+Foxa2-mediated trophic actions.
The effects of astrocyte cografting in this study were dramatic, with almost complete behavioral restoration and extensive DA neuron engraftments cografted for at least 6 months after transplantation in PD rats. As donor cell types and conditions for cell preparation/transplantation are different among studies, the cell therapeutic effects attained by astrocyte cografting in this study cannot be compared with those in other studies. However, the dramatic cell therapeutic outcomes shown in this study, especially long-term DA neuron engraftment, were not comparable to those shown in our previous transplantation data obtained with similar experimental conditions. Specifically, we have recently reported a striking improvement in cell therapeutic efficacy by transplanting rat VM-NPCs engineered with Lin28a (41). Although DA neuron engraftment attained by Lin28a-NPC transplantation was very dramatic during the first 2 months after transplantation, the number of surviving DA neurons in the grafts decreased from 4,523 cells/animal at 2 months to 1,328 cells/animal on average at 6 months after transplantation. In contrast, 3,762 and 3,916 TH+ DA neuronal cells per animal were detected in PD rats by cografting NPCs with VM-Ast and N+F-VM-Ast, respectively (without any modifications to donor NPCs to improve their therapeutic capacity) 6 months after transplantation, strongly indicating the necessity of a host brain modification strategy, especially for long-term donor cell survival and therapeutic efficacy.
In conclusion, we propose astrocyte cografting as a future option in cell therapeutic approaches for PD. In addition, grafting astrocytes alone could exert therapeutic efficacy by improving brain environments, in which remaining endogenous mDA neurons in the SN extend axonal outgrowths to release DA into the striatum and striatal GABAergic interneurons are rescued (79). Astrocytes exert panneuronal trophic actions, in which total neuronal yields, including those of glutamatergic and GABAergic neurons, are also promoted by factors released from cultured astrocytes (Supplemental Figure 9). Thus, the astrocyte cografting strategy could be utilized to treat other CNS disorders. For the clinical application of this therapy in PD, our future studies will address using VM-specific astrocytes and NPCs of human origin in humanized model systems.
-
Methods
Cell cultures
NPC culture. NPCs with DA neurogenic potential were cultured from the VMs of mouse embryos (imprinting control region [ICR]) at E10.5 or Sprague-Dawley rat embryos at E12. The VM-NPCs were expanded in serum-free N2 medium supplemented with the mitogens basic FGF (bFGF) (20 ng/ml; R&D Systems) and epithelial growth factor (EGF) (20 ng/ml; R&D Systems, only for mouse cultures) to reach a confluency of more than 70% (usually for 3–4 days) and passaged. After an additional NPC expansion, cells were harvested for coculture and other experiments or directly induced to differentiate by withdrawing the mitogens (in CM treatment experiments). NPCs were also cultured from the Ctx, a nondopaminergic brain region (mouse at E12 or rat at E14), and used as control cells in the following experiments.
Astrocyte culture. Astrocytes were isolated from mouse or rat VMs or Ctx on P5–P7 and cultured in astro-medium (80). In brief, VMs were removed, triturated in DMEM (Life Technologies) containing 10% FBS (HyClone), and plated in 75-cm2 T-flasks. When cell confluence reached 80%–90%, cells were harvested with 0.1% trypsin and subcultured on a culture surface coated with poly-d-lysine (PDL) (MilliporeSigma). From 4 to 6 days later, microglia were eliminated by shaking at 2 g on an orbital shaker. After culturing for 7 days, astrocytes were harvested for coculture experiments or further cultured for an additional 8 days in N2 to prepare the CM. As described above, minor populations of microglia were present even after the microglia elimination procedure. Astrocyte culture containing a minor microglia population was used in this study. To estimate the effects of minor microglia contamination, microglia-free astrocyte culture was established by treating the cultures with 0.06% trypsin in DMEM/F12 for 20 to 30 minutes after the shaking procedure and discarding floating cells.
Coculture. VM-NPCs with DA neurogenic potential were harvested and mixed with the Ctx-Ast or VM-Ast at a 2:1 ratio of VM-NPCs to astrocytes. The mixed cells were plated, and differentiation of VM-NPCs was directly induced in serum-free N2 medium. In the control cultures, VM-NPCs were mixed with nondopaminergic Ctx-NPCs.
CM treatment
As described above, fresh N2 medium (or HBSS) was added to the astrocyte cultures and CM was collected every other day for 8 days. The control CM was similarly prepared in the Ctx-NPC cultures during 6 days of differentiation. The volume of the CMs was adjusted to 0.1–0.15 mg of protein/ml, filtered at 0.45 μm, and kept at −80°C until use. The CMs were diluted with N2 medium (1:1, v/v) before adding to the cells in culture.
Nurr1+Foxa2 transduction
Lentiviral vectors expressing Nurr1 or Foxa2 under the control of the CMV promoter were generated by inserting the respective cDNA into the multi-cloning site of pCDH (System Biosciences). The empty backbone vector (pCDH) was used as a negative control. The lentiviruses were produced and used for transducing in vitro cultures as described (81). Titers of the lentiviruses were determined using a QuickTiter HIV Lentivirus Quantitation Kit (Cell Biolabs) and 20 ul each of the Nurr1 and Foxa2 viruses with 106 transducing units (TU)/ml were mixed with 2 ml of media and added to 1–1.5 × 106 cells per 6-cm dish for the transduction reaction.
Immunostaining
Cultured cells were fixed with 4% paraformaldehyde (PFA) in PBS and blocked in 0.3% Triton X-100 with 1% BSA for 40 minutes, then incubated with primary antibodies overnight at 4°C. Primary antibody information is shown in Supplemental Table 2. The secondary antibodies used for visualization were as follows: Cy3 (1:200, Jackson Immunoresearch Laboratories) or Alexa Fluor 488 (1:200, Life Technologies). The stained cells were mounted with VECTASHIELD with DAPI mounting solution (Vector Laboratories), and photographs were obtained by an epifluorescence microscope (Leica) and confocal microscope (Leica PCS SP5).
Transplantation and histological procedures
Hemi-parkinsonian was induced in adult female Sprague-Dawley rats (220–250 g) by unilateral stereotactic injection of 3 μl of 6-hydroxydopamine (6-OHDA) (8 μg/μl; MilliporeSigma) into the right side of the substantia nigra (AP –4.8 mm, ML –1.8 mm, V –8.2 mm) and the median forebrain bundle (AP –4.4 mm, ML –1.2 mm, V –7.8 mm). The incisor bar was set at –3.5 mm, and AP and ML coordinates are given relative to bregma. Rats with 300 turns/h ipsilateral to the lesion in an amphetamine-induced rotation test were selected. For transplantation, rat E12 VM-NPCs were expanded and mixed with the Ctx-Ast, VM-Ast, N+F-VM-Ast, or E14 Ctx-NPCs (control) at a 2:1 ratio. Mixed cells (3 μl, 1.5 × 105 cells/μl) were injected over a 10-minute period into each of 2 sites in the striatum (coordinates in AP, ML, and V relative to bregma and dura: [a] 0.07, –0.30, –0.55; [b] –0.10, –0.40, –0.50; incisor bar set at 3.5 mm below zero) under anesthesia induced by Zoletil (100 μl/100 g, 50 mg/ml body weight) mixed with Rompun (23.32 mg/ml). The needle (22 gauge) was left in place for 5 minutes after the completion of each injection. Rats received daily injections of cyclosporine A (10 mg/kg, i.p.) starting 1 day before the grafting and continuing for 1 month and were maintained without the immunosuppressant for the rest of the posttransplantation period. Six months after transplantation, animals were anesthetized and perfused transcardially with 4% PFA. Brains were removed and immersed in 30% sucrose in PBS overnight, frozen in Tissue-Tek (Sakura Finetek), and then sliced on a freezing microtome (Leica). Free-floating brain sections (30 μm thick) were subjected to immunohistochemistry as described above, and images were obtained with a confocal microscope (Leica). In the experiment to examine host environments of the transplanted brains, animals were sacrificed at 1 month after transplantation, grafted brains were sliced at a thickness of 1 mm on a rat brain slice matrix (ZIVIC Instruments), and 8 to 12 regions of the graft-host interface (ca 2 × 2 mm)/graft were dissected and subjected to qPCR analyses. Cells immunoreactive for neurotrophic and proinflammatory glial markers were also counted along the graft-host interfaces of cryosectioned brain slices at 7 to 10 days after transplantation.
Behavior tests
Animal behaviors were assessed using amphetamine-induced rotation, step adjustment, and cylinder tests as previously described (16).
Cell counting and statistical analysis
Immunostained and DAPI-stained cells were counted in 9 to 20 random areas of each culture coverslip using an eyepiece grid at a magnification of ×200 or ×400. For every figure, data are expressed as the mean ± SEM and statistical tests are use as appropriate. Statistical comparisons were made using Student’s t test (unpaired), 2-tailed, or 1-way ANOVA followed by Bonferroni’s post hoc analysis using SPSS (Statistics 21; IBM Inc.). A P value of less than 0.05 was considered significant.
Study approval
All procedures for animal experiments were approved by the IACUC at Hanyang College of Medicine, Seoul, South Korea (approval number 2016-0113A). Experiments were performed in accordance with NIH guidelines.
More information, including a list of PCR primers (Supplemental Table 1), is shown in Supplemental Methods.
-
Author contributions
JJS designed research studies, conducted experiments, acquired data, analyzed data, and wrote the manuscript. SMO designed research studies and conducted experiments. OCK conducted experiments. HSL analyzed data. MYC designed research studies and conducted experiments. NW conducted experiments. EL conducted experiments. WS designed research studies, analyzed data, and wrote the manuscript. SEL conducted experiments. SC designed research studies, analyzed data, and wrote the manuscript. HA conducted experiments. CJL designed research studies, analyzed data, and wrote the manuscript. SHL designed research studies, analyzed data, and wrote the manuscript.
-
Acknowledgments
This work was supported by grants from the Medical Research Center (2017R1A5A2015395, NRF-2017R1A2B2002220, and 2017M3A9B4062401), funded by the National Research Foundation of Korea (NRF) of the Ministry of Science and ICT, South Korea.
Address correspondence to: Sang-Hun Lee, Department of Biochemistry and Molecular Biology, College of Medicine, Hanyang University, 17 Haengdang-dong, Sungdong-gu, Seoul 133-791, South Korea. Phone: 82.2.2220.0625; Email: leesh@hanyang.ac.kr.
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(1):463–482.https://doi.org/10.1172/JCI93924.
See the related Commentary at Creating a graft-friendly environment for stem cells in diseased brains.
-
References
- Goldman SA. Stem and progenitor cell-based therapy of the central nervous system: hopes, hype, and wishful thinking. Cell Stem Cell. 2016;18(2):174–188.
- Steinbeck JA, Studer L. Moving stem cells to the clinic: potential and limitations for brain repair. Neuron. 2015;86(1):187–206.
- Widner H, Brundin P. Immunological aspects of grafting in the mammalian central nervous system. A review and speculative synthesis. Brain Res. 1988;472(3):287–324.View this article via: PubMed Google Scholar
- Barker RA, Widner H. Immune problems in central nervous system cell therapy. NeuroRx. 2004;1(4):472–481.
- Ideguchi M, Shinoyama M, Gomi M, Hayashi H, Hashimoto N, Takahashi J. Immune or inflammatory response by the host brain suppresses neuronal differentiation of transplanted ES cell-derived neural precursor cells. J Neurosci Res. 2008;86(9):1936–1943.
- Horner PJ, Palmer TD. New roles for astrocytes: the nightlife of an ‘astrocyte’. La vida loca! Trends Neurosci. 2003;26(11):597–603.
- Nedergaard M, Ransom B, Goldman SA. New roles for astrocytes: redefining the functional architecture of the brain. Trends Neurosci. 2003;26(10):523–530.
- Molofsky AV, et al. Astrocytes and disease: a neurodevelopmental perspective. Genes Dev. 2012;26(9):891–907.
- Hamby ME, Sofroniew MV. Reactive astrocytes as therapeutic targets for CNS disorders. Neurotherapeutics. 2010;7(4):494–506.
- Bronzuoli MR, Iacomino A, Steardo L, Scuderi C. Targeting neuroinflammation in Alzheimer’s disease. J Inflamm Res. 2016;9:199–208.
- Sofroniew MV, Vinters HV. Astrocytes: biology and pathology. Acta Neuropathol. 2010;119(1):7–35.
- Neumann H, Kotter MR, Franklin RJ. Debris clearance by microglia: an essential link between degeneration and regeneration. Brain. 2009;132(Pt 2):288–295.View this article via: PubMed Google Scholar
- Liddelow SA, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541(7638):481–487.
- Ransohoff RM. A polarizing question: do M1 and M2 microglia exist? Nat Neurosci. 2016;19(8):987–991.
- Saijo K, et al. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009;137(1):47–59.
- Oh SM, et al. Combined Nurr1 and Foxa2 roles in the therapy of Parkinson’s disease. EMBO Mol Med. 2015;7(5):510–525.
- Zhang Y, Barres BA. Astrocyte heterogeneity: an underappreciated topic in neurobiology. Curr Opin Neurobiol. 2010;20(5):588–594.
- Hochstim C, Deneen B, Lukaszewicz A, Zhou Q, Anderson DJ. Identification of positionally distinct astrocyte subtypes whose identities are specified by a homeodomain code. Cell. 2008;133(3):510–522.
- Emsley JG, Macklis JD. Astroglial heterogeneity closely reflects the neuronal-defined anatomy of the adult murine CNS. Neuron Glia Biol. 2006;2(3):175–186.
- Yeh TH, Lee DY, Gianino SM, Gutmann DH. Microarray analyses reveal regional astrocyte heterogeneity with implications for neurofibromatosis type 1 (NF1)-regulated glial proliferation. Glia. 2009;57(11):1239–1249.
- Hall AC, Mira H, Wagner J, Arenas E. Region-specific effects of glia on neuronal induction and differentiation with a focus on dopaminergic neurons. Glia. 2003;43(1):47–51.
- Hofmann MA, et al. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97(7):889–901.
- Kressin K, Kuprijanova E, Jabs R, Seifert G, Steinhäuser C. Developmental regulation of Na+ and K+ conductances in glial cells of mouse hippocampal brain slices. Glia. 1995;15(2):173–187.
- Hwang EM, et al. A disulphide-linked heterodimer of TWIK-1 and TREK-1 mediates passive conductance in astrocytes. Nat Commun. 2014;5:3227. View this article via: PubMed Google Scholar
- Filous AR, Miller JH, Coulson-Thomas YM, Horn KP, Alilain WJ, Silver J. Immature astrocytes promote CNS axonal regeneration when combined with chondroitinase ABC. Dev Neurobiol. 2010;70(12):826–841.
- Haas C, Fischer I. Human astrocytes derived from glial restricted progenitors support regeneration of the injured spinal cord. J Neurotrauma. 2013;30(12):1035–1052.
- Noble M, Davies JE, Mayer-Pröschel M, Pröschel C, Davies SJ. Precursor cell biology and the development of astrocyte transplantation therapies: lessons from spinal cord injury. Neurotherapeutics. 2011;8(4):677–693.
- Smith GM, Miller RH. Immature type-1 astrocytes suppress glial scar formation, are motile and interact with blood vessels. Brain Res. 1991;543(1):111–122.
- Smith GM, Silver J. Transplantation of immature and mature astrocytes and their effect on scar formation in the lesioned central nervous system. Prog Brain Res. 1988;78:353–361.
- Volpicelli F, De Gregorio R, Pulcrano S, Perrone-Capano C, di Porzio U, Bellenchi GC. Direct regulation of Pitx3 expression by Nurr1 in culture and in developing mouse midbrain. PLoS ONE. 2012;7(2):e30661.
- Watakabe A, et al. Comparative analysis of layer-specific genes in Mammalian neocortex. Cereb Cortex. 2007;17(8):1918–1933.
- Peña de Ortiz S, Maldonado-Vlaar CS, Carrasquillo Y. Hippocampal expression of the orphan nuclear receptor gene hzf-3/nurr1 during spatial discrimination learning. Neurobiol Learn Mem. 2000;74(2):161–178.
- Sommer L, Rao M. Neural stem cells and regulation of cell number. Prog Neurobiol. 2002;66(1):1–18.
- Rhee YH, et al. Neural stem cells secrete factors facilitating brain regeneration upon constitutive Raf-Erk activation. Sci Rep. 2016;6:32025. View this article via: PubMed Google Scholar
- Roeper J. Dissecting the diversity of midbrain dopamine neurons. Trends Neurosci. 2013;36(6):336–342.
- Dong J, Li S, Mo JL, Cai HB, Le WD. Nurr1-based therapies for Parkinson’s disease. CNS Neurosci Ther. 2016;22(5):351–359.
- Blaudin de Thé FX, Rekaik H, Prochiantz A, Fuchs J, Joshi RL. Neuroprotective transcription factors in animal models of Parkinson disease. Neural Plast. 2016;2016:6097107. View this article via: PubMed Google Scholar
- Decressac M, Volakakis N, Björklund A, Perlmann T. NURR1 in Parkinson disease--from pathogenesis to therapeutic potential. Nat Rev Neurol. 2013;9(11):629–636.
- Arenas E. Foxa2: the rise and fall of dopamine neurons. Cell Stem Cell. 2008;2(2):110–112.
- Decressac M, Kadkhodaei B, Mattsson B, Laguna A, Perlmann T, Björklund A. α-Synuclein-induced down-regulation of Nurr1 disrupts GDNF signaling in nigral dopamine neurons. Sci Transl Med. 2012;4(163):163ra156.
- Rhee YH, et al. LIN28A enhances the therapeutic potential of cultured neural stem cells in a Parkinson’s disease model. Brain. 2016;139(Pt 10):2722–2739.View this article via: PubMed Google Scholar
- Hof PR, Morrison JH. The aging brain: morphomolecular senescence of cortical circuits. Trends Neurosci. 2004;27(10):607–613.
- Trendelenburg G, Dirnagl U. Neuroprotective role of astrocytes in cerebral ischemia: focus on ischemic preconditioning. Glia. 2005;50(4):307–320.
- Sandhu JK, et al. Astrocyte-secreted GDNF and glutathione antioxidant system protect neurons against 6OHDA cytotoxicity. Neurobiol Dis. 2009;33(3):405–414.
- Chung S, et al. Wnt1-lmx1a forms a novel autoregulatory loop and controls midbrain dopaminergic differentiation synergistically with the SHH-FoxA2 pathway. Cell Stem Cell. 2009;5(6):646–658.
- Gonzalez-Reyes LE, et al. Sonic hedgehog maintains cellular and neurochemical homeostasis in the adult nigrostriatal circuit. Neuron. 2012;75(2):306–319.
- Ye W, Shimamura K, Rubenstein JL, Hynes MA, Rosenthal A. FGF and Shh signals control dopaminergic and serotonergic cell fate in the anterior neural plate. Cell. 1998;93(5):755–766.
- Lee SH, Lumelsky N, Studer L, Auerbach JM, McKay RD. Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nat Biotechnol. 2000;18(6):675–679.
- Kriks S, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480(7378):547–551.View this article via: PubMed Google Scholar
- Castelo-Branco G, Sousa KM, Bryja V, Pinto L, Wagner J, Arenas E. Ventral midbrain glia express region-specific transcription factors and regulate dopaminergic neurogenesis through Wnt-5a secretion. Mol Cell Neurosci. 2006;31(2):251–262.
- Panchision DM, et al. An immortalized, type-1 astrocyte of mesencephalic origin source of a dopaminergic neurotrophic factor. J Mol Neurosci. 1998;11(3):209–221.
- Hao HX, et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485(7397):195–200.
- Christopherson KS, et al. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120(3):421–433.
- Allen NJ, et al. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature. 2012;486(7403):410–414.View this article via: PubMed Google Scholar
- Chen S, Lewallen M, Xie T. Adhesion in the stem cell niche: biological roles and regulation. Development. 2013;140(2):255–265.
- Rozo M, Li L, Fan CM. Targeting β1-integrin signaling enhances regeneration in aged and dystrophic muscle in mice. Nat Med. 2016;22(8):889–896.
- Lukjanenko L, et al. Loss of fibronectin from the aged stem cell niche affects the regenerative capacity of skeletal muscle in mice. Nat Med. 2016;22(8):897–905.
- Nishimura K, et al. Estradiol facilitates functional integration of iPSC-derived dopaminergic neurons into striatal neuronal circuits via activation of integrin α5β1. Stem Cell Reports. 2016;6(4):511–524.
- Marchionini DM, Collier TJ, Camargo M, McGuire S, Pitzer M, Sortwell CE. Interference with anoikis-induced cell death of dopamine neurons: implications for augmenting embryonic graft survival in a rat model of Parkinson’s disease. J Comp Neurol. 2003;464(2):172–179.
- Siebert JR, Conta Steencken A, Osterhout DJ. Chondroitin sulfate proteoglycans in the nervous system: inhibitors to repair. Biomed Res Int. 2014;2014:845323. View this article via: PubMed Google Scholar
- McKerracher L, David S, Jackson DL, Kottis V, Dunn RJ, Braun PE. Identification of myelin-associated glycoprotein as a major myelin-derived inhibitor of neurite growth. Neuron. 1994;13(4):805–811.
- Bittner S, Afzali AM, Wiendl H, Meuth SG. Myelin oligodendrocyte glycoprotein (MOG35-55) induced experimental autoimmune encephalomyelitis (EAE) in C57BL/6 mice. J Vis Exp. 2014;(86):51275. View this article via: PubMed Google Scholar
- Kizil C, Kyritsis N, Brand M. Effects of inflammation on stem cells: together they strive? EMBO Rep. 2015;16(4):416–426.
- Chapman KZ, et al. Inflammation without neuronal death triggers striatal neurogenesis comparable to stroke. Neurobiol Dis. 2015;83:1–15.
- Barkho BZ, et al. Identification of astrocyte-expressed factors that modulate neural stem/progenitor cell differentiation. Stem Cells Dev. 2006;15(3):407–421.
- Marchetti B, et al. Uncovering novel actors in astrocyte-neuron crosstalk in Parkinson’s disease: the Wnt/β-catenin signaling cascade as the common final pathway for neuroprotection and self-repair. Eur J Neurosci. 2013;37(10):1550–1563.
- Rothhammer V, et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med. 2016;22(6):586–597.
- Teige I, Liu Y, Issazadeh-Navikas S. IFN-beta inhibits T cell activation capacity of central nervous system APCs. J Immunol. 2006;177(6):3542–3553.
- Ejlerskov P, et al. Lack of neuronal IFN-β-IFNAR causes Lewy body- and Parkinson’s disease-like dementia. Cell. 2015;163(2):324–339.
- Olsen ML, Khakh BS, Skatchkov SN, Zhou M, Lee CJ, Rouach N. New insights on astrocyte ion channels: critical for homeostasis and neuron-glia signaling. J Neurosci. 2015;35(41):13827–13835.
- Hansson E, et al. Astroglia and glutamate in physiology and pathology: aspects on glutamate transport, glutamate-induced cell swelling and gap-junction communication. Neurochem Int. 2000;37(2-3):317–329.
- Hu X, et al. Microglia/macrophage polarization dynamics reveal novel mechanism of injury expansion after focal cerebral ischemia. Stroke. 2012;43(11):3063–3070.
- Eddleston M, Mucke L. Molecular profile of reactive astrocytes--implications for their role in neurologic disease. Neuroscience. 1993;54(1):15–36.
- Pekny M, Pekna M. Astrocyte reactivity and reactive astrogliosis: costs and benefits. Physiol Rev. 2014;94(4):1077–1098.
- Boche D, Perry VH, Nicoll JA. Review: activation patterns of microglia and their identification in the human brain. Neuropathol Appl Neurobiol. 2013;39(1):3–18.
- Nishimura K, Murayama S, Takahashi J. Identification of neurexophilin 3 as a novel supportive factor for survival of induced pluripotent stem cell-derived dopaminergic progenitors. Stem Cells Transl Med. 2015;4(8):932–944.
- Chen C, Chan A, Wen H, Chung SH, Deng W, Jiang P. Stem and progenitor cell-derived astroglia therapies for neurological diseases. Trends Mol Med. 2015;21(11):715–729.
- Chen SH, et al. Microglial regulation of immunological and neuroprotective functions of astroglia. Glia. 2015;63(1):118–131.
- Proschel C, Stripay JL, Shih CH, Munger JC, Noble MD. Delayed transplantation of precursor cell-derived astrocytes provides multiple benefits in a rat model of Parkinsons. EMBO Mol Med. 2014;6(4):504–518.
- Heinrich C, et al. Generation of subtype-specific neurons from postnatal astroglia of the mouse cerebral cortex. Nat Protoc. 2011;6(2):214–228.
- Yi SH, et al. Foxa2 acts as a co-activator potentiating expression of the Nurr1-induced DA phenotype via epigenetic regulation. Development. 2014;141(4):761–772.
-
Version history
- Version 1 (December 11, 2017): Electronic publication
- Version 2 (December 18, 2017): Noviana Wulansari's surname corrected in author list
- Version 3 (January 2, 2018): Print issue publication
- Version 4 (May 25, 2021): Affiliation information corrected for HA and CJL.



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)










