Advertisement
Review Series Free access | 10.1172/JCI37934
The genetic and neurobiologic compass points toward common signaling dysfunctions in autism spectrum disorders
Pat Levitt and Daniel B. Campbell
Vanderbilt Kennedy Center for Research on Human Development and Department of Pharmacology, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Address correspondence to: Pat Levitt, Zilkha Neurogenetic Institute, Keck School of Medicine of University of Southern California, 1501 San Pablo Street, Los Angeles, California 90087, USA. Phone: (323) 442-1509; Fax: (323) 442-2145; E-mail: plevitt@usc.edu.
Find articles by Levitt, P. in: PubMed | Google Scholar
Vanderbilt Kennedy Center for Research on Human Development and Department of Pharmacology, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Address correspondence to: Pat Levitt, Zilkha Neurogenetic Institute, Keck School of Medicine of University of Southern California, 1501 San Pablo Street, Los Angeles, California 90087, USA. Phone: (323) 442-1509; Fax: (323) 442-2145; E-mail: plevitt@usc.edu.
Find articles by Campbell, D. in: PubMed | Google Scholar
Published April 1, 2009 - More info
J Clin Invest. 2009;119(4):747–754. https://doi.org/10.1172/JCI37934.
© 2009 The American Society for Clinical Investigation
-
Abstract
Autism spectrum disorder (ASD) is a common neurodevelopmental disorder with high heritability. Here, we discuss data supporting the view that there are at least two distinct genetic etiologies for ASD: rare, private (de novo) single gene mutations that may have a large effect in causing ASD; and inherited, common functional variants of a combination of genes, each having a small to moderate effect in increasing ASD risk. It also is possible that a combination of the two mechanisms may occur in some individuals with ASD. We further discuss evidence from individuals with a number of different neurodevelopmental syndromes, in which there is a high prevalence of ASD, that some private mutations and common variants converge on dysfunctional ERK and PI3K signaling, which negatively impacts neurodevelopmental events regulated by some receptor tyrosine kinases.
Autism spectrum disorder (ASD) is a syndrome characterized by a triad of core deficits: disturbances in social behavior, atypical verbal and nonverbal communication, and restricted interests that can be accompanied by repetitive behavior. The clinical diagnosis, which includes individuals with any one of a spectrum of neurodevelopmental conditions (including autism, Rett syndrome, pervasive developmental disorder–not otherwise specified, and Asperger syndrome), is made in 1 of every 150 individuals and is four times more prevalent in boys than girls (1). While ASD is among the most heritable psychiatric disorders defined in the Diagnostic and statistical manual of mental disorders (4th edition) (2), it is not a static or simple disorder with fixed effects on a circumscribed age. Instead, equally fundamental facets of pathology emerge at different points of maturation of the child. Moreover, the disorder does not result in immutable social and cognitive deficits, but rather the core symptoms typically change over time and to different degrees. Co-occurring medical conditions (sleep problems, epilepsy, and gastrointestinal symptoms) and psychiatric disturbances (anxiety, obsessive-compulsive disorder, and aggression) are common and can appear at different ages in children on the spectrum.
Contemporary hypotheses of the causes of ASD often include experience-dependent processes through which atypical gene-by-environment (G X E) interactions yield pathophysiology in later emerging systems that underlie social and communication competencies. The later emergence of symptoms is consistent with the concept that developmental differentiation, whether at the cellular, circuit, or systems level, occurs from the bottom up; behavior develops from basic sensory and perceptual systems that feed into higher integration centers (3–5). Impairments in initial basic processes become expressed in ever more complex systems, with the population heterogeneity of the clinical features of ASD expected to increase from infancy to childhood and through adolescence. However, it is not clear whether the factors that contribute to the developmental diversification and phenotypic heterogeneity of ASD are related to a complex genetic etiology of ASD itself or whether they also involve the interaction between ASD-specific and -nonspecific functional features.
Clinical researchers have noted the importance of addressing disorder heterogeneity in the study of ASD (6–8). In this regard, the conundrum facing investigators is connecting the well-defined, highly heritable nature of ASD with the striking differences in the initial expression of core symptoms, progressive changes over time, and differential response to interventions. Thus, a major goal of the current interdisciplinary research agenda is not only to explain the etiologies of ASD but also to understand the syndrome-specific and -nonspecific factors that influence variability in the relative risk of developing ASD, in the developmental course of symptom presentation, in the responsiveness to treatment, and in the co-occurrence of other medical dysfunctions (6, 8–11). This Review highlights the current understanding of ASD genetics, key pathophysiological findings from behavior and imaging studies, and potential G X E interactions that may be at the core of ASD expression. The Review ends with what we believe to be a novel hypothesis that combines recent genetic findings to propose one potential mechanism of heterogeneity in ASD.
-
Distinct genetic mechanisms can result in ASD
Based on studies in mono- and dizygotic twins (12, 13), the estimated heritability of ASD is approximately 0.90. This far exceeds the estimated heritability of other common polygenic diseases, including cancer, heart disease, schizophrenia, and depression. The focus, therefore, on defining the underlying genetic etiology of ASD has escalated dramatically during the current decade, in parallel with the rapid development of affordable genomic methods that have facilitated the analysis of large populations and entire genomes. Like other complex disorders, however, the most critical challenges of the field lie in defining the heritability of risk for developing ASD that may be due to G X E factors that alter the trajectory of brain development and the direct impact of de novo or heritable gene variation on brain development. ASD is a spectrum of disorders, in which there are differences in the degree of severity of the three core symptoms as well as other co-occurring mental health and physical conditions. Thus, to emphasize the functional importance of recognizing many different kinds of ASD, the term “autisms” has been used (14). The behaviors that are disrupted in ASD are complex and develop through a bottom-up assembly of simple to more complex brain circuits that control very basic processes such as physiological homeostasis and more complex tasks such as being motivated to pay attention to certain cues in the environment that regulate outward social behavior and verbal and nonverbal communication. The heterogeneity of ASD is entirely consistent with the concept that different genetic mechanisms may influence brain circuit development at different levels of the hierarchy (5). The field thus is moving away from defining the ASD genes to defining unique phenotypic features of stratified populations of children, adolescents, and adults that may relate to specific genetic etiologies, such as increased risk due to common allelic variations, rare mutations, or copy number variation (CNV) (Figure 1). Implicit in this view is that there will not be identification of genetic risks that map one-to-one with behavioral dysfunction; that is, while there are genetic variants that are enriched in populations with particular dysfunctions, such as language, there are no genes that directly regulate social behavior or language. Instead, genetic vulnerability resides in the disruption of cellular processes, due to the disruption of proteins encoded by genes, in specific brain circuits that may also be influenced by G X E mechanisms. Research findings emerging from human genetic and animal studies suggest that disruption of a key developmental process, synapse formation and stabilization (synaptogenesis), is a final common path in ASD etiology. Different molecular mechanisms may contribute to increasing ASD risk, including disturbances in the assembly of structural proteins needed to build synapses, such as the neuroligins and neurexins, and dysfunctional cellular signaling pathways that control synaptogenesis.
 Figure 1
Figure 1Current experimental approaches to determining genetic etiologies for ASD. These approaches include whole-genome analyses that identify disorder-related sequences or CNVs in genes that exhibit preferential inheritance patterns or de novo appearance in individuals with ASD. The current challenges include the translation of these genetic findings to define the biological consequences of the variations, to determine the influence on defined clinical phenotypes of ASD, and eventually to design new intervention strategies.
-
Distinct patterns of heritability of risk alleles in ASD
As noted above, ASD is highly heritable, and current studies suggest that there are multiple mechanisms through which different types of gene mutations increase risk of developing the disorder (15–17). There are a number of considerations that are key to successful genetic studies of ASD. First, because of the heterogeneity of the disorder, it is necessary to analyze large numbers of individuals with ASD. Second, each individual gene is likely to have very small effects on disease risk, but in combination with other genes and/or G X E factors, an individual gene may encode a protein that functions in a key cellular process, which, when disrupted, contributes to disease pathophysiology. Third, disorder emergence through de novo genetic mutations or heritability of gene-specific functional polymorphisms in the DNA sequence transmitted from parent to child may underlie distinct but equivalently valid ASD etiologies. Fourth, distinct genetic etiologies, together with different environmental factors, may be part of ASD heterogeneity. Last, the nature of the core behavioral dimensions that characterize ASD emerge through perturbation of developing brain circuits. Disruption at distinct levels of the organizational and functional hierarchy relate to the heterogeneity in social behavior and communication capabilities.
The aim of genetic studies of ASD should be to identify functional variants that contribute to ASD risk. A thorough recent review provides a detailed listing of up-to-date genetic findings (16). One approach with great promise for the identification of candidate genes and pathways is analysis of CNV (see The basics of CNVs) (18, 19). However, as with single gene mutations and common variants, CNV analyses need to be interpreted with extreme caution for a number of reasons. First, the presence of a de novo CNV in an individual with ASD does not necessarily imply it is associated with increased risk of developing the disorder. Further, CNVs typically are not fully penetrant, meaning that they may be present in individuals who do not have an ASD. CNVs were first described in healthy control individuals, with more than 11 CNVs per individual (20), indicating that having multiple CNVs is not pathologic. Only formal genetic association analyses involving large sample sizes should be used to imply a particular CNV is associated with disorder risk. Second, the presence of a CNV does not necessarily imply functional disruption. Analyses of CNVs in the human adult cerebral cortex indicate that more than 50% of mature neurons are aneuploid (21, 22), and experiments in mice indicate that CNVs in cortical neurons may have little impact on function (23). Further, germline deletion of both copies of certain genes in experimental animals can result in mutants without a detectable phenotype. This suggests that due to adaptive processes, gene dosage in the form of CNV does not lead necessarily to dramatic functional changes in vivo. Third, CNV in peripheral blood cells, the cells typically analyzed in humans, may not relate in a one-to-one fashion to CNVs in neurons. Indeed, the number of CNVs in the human cerebral cortex is approximately 7-fold higher than in peripheral blood cells (21), and thus, analysis of peripheral blood may identify some, but not necessarily all, of the CNVs occurring in neurons that contribute to ASD-related disturbances of brain architecture and circuitry. Fourth, de novo CNVs are observed in 7%–10% of cases from simplex families (families with only one child with ASD), 2%–3% of cases from multiplex families (families with more than one child with ASD), and 1% of controls (24, 25). The de novo CNVs that occur in a subset of individuals with ASD in multiplex families may influence the severity of the disorder, rather than contributing directly to the expression of the disorder. Despite these cautions, CNV analysis can be used to identify candidate genes that can be tested further for functional effects that may contribute to ASD susceptibility (18, 19).
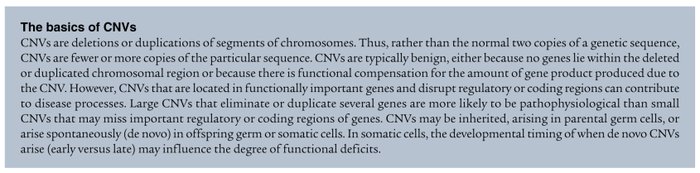
We are beginning to recognize that inheritance of rare or common functional alleles is only one genetic mechanism that increases disorder risk. Private (de novo) functional mutations also impart genetic risk. Analyses suggest that, at the genetic and behavioral levels, multiplex families may be fundamentally different from simplex families (24, 26–29). Furthermore, multiple genes or even multiple mutations of the same gene may be involved in the etiology of the same clinically diagnosed disorder in different individuals. For example, there are over 50 genes that carry mutations known to cause nonsyndromic retinitis pigmentosa (30). Conversely, there are more than 130 distinct catalogued mutations of the 7-dehydrocholesterol reductase (DHCR7) gene in individuals with the monogenic disorder Smith-Lemli-Opitz syndrome (31). Heritable, high-risk mutations in breast cancer, such as those in the breast cancer 1, early onset (BRCA1) gene, are balanced by more common variants in multiple genes discovered through whole-genome association studies (WGASs). Given the range of possibilities for disorder etiology, sample numbers are clearly important. For example, WGASs have examined between 500,000 and 1,000,000 SNPs simultaneously in thousands of patient samples for diabetes and coronary artery disease (18, 32–34). These diseases, with arguably less complex pathophysiology than ASD, only recently have had the sample power to generate statistically reliable data that reveal common SNPs with disease-related heritability patterns in multiple genes. Similarly, rare functional mutations in ASD candidate risk genes initially may seem to be overrepresented in the clinical population compared with unrelated controls, but recent analysis demonstrates that even these types of studies more accurately reflect clinical findings when larger sample populations are assessed (35).
-
Syndromic disorders and rare mutations point the way
Although not understood from an etiological or pathophysiological perspective, it is now clear that rare neurodevelopmental disorders (<1 in 10,000) are becoming increasingly important to study in greater detail because of their relationship to ASD. Higher penetrance of ASD diagnosis (far greater than the 0.75% observed in the general population) is reported in children who have genetically diverse neurodevelopmental syndromic disorders, including Angelman syndrome, Fragile X syndrome (FraX), Rett syndrome, Smith-Lemli-Opitz syndrome, Timothy syndrome, neurofibromatosis, and tuberous sclerosis. It is important to emphasize that each syndrome is characterized by fundamentally different gene mutations, which presumably impart distinct molecular pathophysiologies (Table 1). However, there are few studies that examine closely the similarities and differences in phenotypic characteristics between single gene (syndromic) and multigenic (idiopathic) ASD (36). The neurodevelopmental syndromic disorders listed in Table 1 are characterized in part by intellectual disability (ID; formally termed mental retardation). A large minority (25%–40%) of individuals with ASD has ID, but ASD is not synonymous with ID. A recent structural MRI study suggests that individuals with FraX, with or without ASD diagnosis, are more closely related in the context of the size of brain structures than those with idiopathic ASD (37). Microarray analysis of lymphocytes from patients with FraX, chromosome 15q deletion, or idiopathic ASD reveal unique patterns of gene expression that may serve as a signature for each disorder, but with a potentially important small subset of overlapping changes in mRNA expression (38). Moreover, detailed neuropathological studies are lacking to compare these syndromes and idiopathic ASD. Although there is likely to be diversity in the pathological targets in each syndrome, there are suggestions of some commonalities. The triad of overlapping dysfunctions (social behavior, communication, and repetitive behavior) across ASD and the syndromes, together with the known brain neuropathology of some of the syndromes, suggests that later neurodevelopmental events, such as synapse formation and maturation, dendritic growth, and myelination, are probably most vulnerable. In addition to the evidence from syndromic disorders, the focus on later events in this Review is supported by the discovery of rare mutations in certain genes that regulate synaptogenesis. A substantial focus has been on the adhesive and structural elements needed for synapse formation, stability, and physiologic maturation. In ASD cases, rare mutations and CNVs have been identified in genes encoding neuroligins, neurexins, contactin-associated protein-2 (CNTNAP2), and SH3 and multiple ankyrin repeat domains 3 (SHANK3). The disruptions are likely to occur in shared forebrain and cerebral cortical circuits. Thus, while not identical to idiopathic ASD, biological and behavioral analyses of syndromic disorders and rare mutations provide a sound approach to discern potential overlapping molecular and brain targets (Table 1).
-
Intracellular kinase signaling in ASD-associated disorders
The potential contribution of defects in adhesion and structural proteins that build synapses to the etiology of ASD has been the subject of many reviews of ASD (16, 17, 39–41). In this Review, we suggest that some neurodevelopmental syndromic disorders and rare mutations point to an additional set of molecular targets. Thus, recognizing that we are attempting to resolve a spectrum of disorders that will not have a single, underlying etiology, findings from studies of certain syndromic disorders with high penetrance of ASD converge on the ERK and PI3K intracellular signaling pathways that we believe deserve increased scrutiny in all forms of ASD. ERK and PI3K activate mammalian target of rapamycin (mTOR), which through other kinases will increase mRNA translation to influence developmental functions as diverse as the cell cycle, cell survival, differentiation, and motility. Receptor tyrosine kinases (RTKs) can signal through either of these intracellular kinase pathways, with cell type and cellular milieu defining the intracellular response (Figure 2). Table 1 reports several syndromic disorders with high penetrance of ASD that involve a primary disruption in signaling through these pathways specifically and others that would disrupt RTK signaling, the primary membrane receptor class that transduces signals through ERK and PI3K. The most convincing connections between ERK/PI3K signaling disruption and ASD are evident in tuberous sclerosis and neurofibromatosis type 1, in which different elements of the ERK/PI3K pathway are disrupted genetically, leading to enhanced mTOR downstream activation (Figure 2). In addition, ERK and PI3K signaling is dependent in part on normal cholesterol biosynthesis, which is absent in Smith-Lemli-Opitz syndrome. For example, Ras signaling, a key upstream mediator of ERK activation, requires cholesterolization. Rare gene mutations of another element of the PI3K signaling pathway, phosphatase and tensin homolog (PTEN), are associated with high prevalence of ASD. Rett syndrome disrupts the X-linked methyl CpG binding protein 2 (MECP2) gene, which encodes a protein that binds to specific regulatory regions of certain genes (based on DNA methylation patterns) that control gene transcription. Methylation status and/or MECP2 binding directly regulates transcription of key genes involved in met proto-oncogene–RTK signaling (MET RTK signaling; MET is also known as HGFR), which our laboratory has implicated in ASD risk (see below). Those genes include those encoding MET, the MET coreceptor CD44, the MET transcriptional regulator SP1, and several proteins in the ERK/PI3K downstream signaling pathway (42).
 Figure 2
Figure 2The MET RTK signaling pathway and genes implicated in ASD risk. Intracellular signaling of MET and other RTKs occurs via the PI3K or ERK1/2 pathways. Rare mutations and CNVs (which are both designated by ‡) or associated common alleles (which are designated by *) have been identified in individuals with ASD in seven genes encoding proteins involved in these signaling pathways. Of note, an association between common MET variants and ASD has been reported for five independent family cohorts. PLAUR and SERPINE1 associations with ASD have been determined in single, large family cohorts (>600 families). Ras disruption in Smith-Lemli-Opitz syndrome is due to alterations in cholesterol biosynthesis (which is designated by †). Also depicted are other proteins that interact with the MET signaling pathway, such as semaphorins, plexins, and other RTKs. MET can signal via the PI3K and the ERK pathway. RTKs, including MET, are involved in key neurodevelopmental processes, including axon guidance, synapse formation, and plasticity. Convergence of many different genetic etiologies suggests that risk via ERK/PI3K signaling may be common in ASD. Risk, severity of the pathophysiology (i.e., intellectual disability), and disorder heterogeneity may relate to differences in genetic and epigenetic points of entry to the pathways. Thus, the impact due to genetic risk, via regulators of ligand availability or RTKs such as MET, may be less severe than the more severe clinical impact (i.e., intellectual disability) from disruption downstream along the intracellular signaling pathways. c-cbl, E3 ubiquitin-protein ligase c-Cbl; rheb, Ras homolog enriched in brain; RSK, ribosomal S6 kinase; uPA, urokinase plasminogen activator.
The various neurodevelopmental syndromic disorders and rare mutations described thus far along the ERK/PI3K pathways result in an increased state of activation of mTOR (Figure 2). Additional evidence for involvement of these intracellular kinase pathways in ASD comes from recent treatment studies in genetically engineered mice that exhibit behavioral and neuropathologic phenotypes that are common in the human neurodevelopmental syndromic disorders. For example, systemic administration of drugs that reduce mTOR activation, such as rapamycin, wortmannin, and RAD001, can reverse behavioral and structural pathology in mice with Pten (43), tuberous sclerosis 1 (Tsc1) (44, 45), and neurofibromin 1 (Nf1) (46, 47) mutations, with no reported side effects.
ASD etiologies also are likely to include environmental factors that work together with genetic risk to drive neurodevelopment systems over the threshold for disorder expression (Figure 3). We therefore hypothesize that different genetic routes to altered RTK function, by way of modulation of ERK/PI3K signaling pathways, combine with environmental factors, such as biochemical stressors, that also modulate these signaling pathways. The G X E interactions either modulate the degree of dysfunction of the core clinical features of ASD or have an impact on neurobiological circuits that are at greater risk for dysfunction, because genetic vulnerability pushes the system closer to disorder threshold.
 Figure 3
Figure 3Contributions of the PI3K pathway to ASD risk threshold. The degree of genetic risk is indicated by shading, with darker color indicating increased risk. The model presents common functional variants in the MET, PLAUR, and SERPINE1 genes that, along with other genetic risk alleles, contribute to risk of developing ASD. Adaptive processes may prevent presentation of ASD, but additional environmental factors or the presence of multiple risk alleles result in idiopathic (multiple genes, each having a small effect) ASD. Mutations further down the PI3K pathway result in syndromic disorders, with penetrance and phenotype severity determined by a decreasing availability of adaptive processes.
Given that ERK/PI3K signaling is widely distributed throughout multiple organ systems, where does disorder specificity arise? One way to think about the issue of specificity is to recognize that signaling through ERK/PI3K is highly influenced by cell type and timing of activation of the RTK signaling systems. For example, there is a potential dichotomy in the molecular mechanisms of ASD and cancer that would involve different genetic risk factors affecting ERK/PI3K signaling. Unequivocal evidence implicates hyperactivated PI3K signaling in a number of malignant cancer types (48–50). In contrast, decreased PI3K activation may contribute in some instances to ASD (26, 51, 52). We are unaware of any studies of cancer frequencies in individuals with ASD. However, disruption of PI3K signaling also has been implicated in other psychiatric disorders of neurodevelopmental origin, such as schizophrenia (53, 54). An altered incidence of various cancers in individuals with schizophrenia is debated (55, 56), but reduced cancer incidence is observed consistently in parents and siblings of individuals with schizophrenia compared with the general population (57–59). These data support the plausibility of a genetic impact, through different mutations or common variants, that increases risk for a neurodevelopmental disorder and decreases cancer risk, a hypothesis that can be tested by epidemiological studies and complete sequencing of candidate genes to identify mutations associated with specific disorders.
One testable facet of our hypothesis is that risk for more global neurodevelopmental disruptions increases when the genetic hits are downstream from the molecular components that are involved in initial RTK activation, which are the growth factors or receptors themselves. Consistent with this, mutations in NF1, AKT, TSC1, and TSC2 typically result in widespread and severe clinical problems such as mild to severe intellectual disabilities, seizure disorder, sensory-motor deficits, and medical dysfunctions (Figure 3). The corollary to this would be that mutations in upstream genes encoding RTKs or proteins that regulate growth factor availability would place signaling through this pathway at risk but require additional genetic and environmental insults to cause neurodevelopmental disruption. Disruption of the development of specific brain circuits would occur, because, unlike their intracellular mediators, upstream signaling elements are not distributed uniformly. Rather, RTKs and growth factors may be concentrated in developing circuits at key periods of development that mediate the maturation of connections underlying specific functions. This hypothesis is consistent with the identification of neuregulin 1 (the ligand for the RTK ERBB4) as a factor for schizophrenia susceptibility (60) and the RTK MET as a factor for ASD risk (26, 27, 52). Although both ERBB4 and MET activate PI3K signaling, the differential timing and patterns of expression of each of these RTKs in developing cerebral cortex (61) may account for the distinct neurodevelopmental disruptions characteristic of each disorder. We have shown that MET is enriched in neocortex, amygdala, septum, and cerebellum, regions implicated in ASD (62).
-
MET in PI3K signaling and ASD
Our own genetic and neuropathological studies of ASD have focused on one of the upstream activators of both ERK and PI3K in various cell types, MET. Much is known about its role in PI3K signaling. Specifically, HGF activation of MET causes phosphorylation of AKT that can be blocked by the PI3K inhibitors LY294002 and wortmannin (63–65). Most relevant to the current discussion, MET activation of PI3K signaling has been demonstrated in neuronal cells, resulting in neuroprotection of cerebellar granule cells (63), cell motility in striatal progenitor cells (66), and protection of cortical neurons from hypoxia-induced insult (67).
It is again important to emphasize that the phenotypic heterogeneity of ASD makes it unlikely that any individual gene will contribute to more than a subset of cases. This creates a natural tension in the field that is attempting to translate genetic findings into plausible biological models of ASD. Thus, there currently is legitimate skepticism regarding any particular candidate gene (16, 17). However, both convergent neurobiological and genetic evidence is emerging to suggest ASD vulnerability may lie, in part, in the well-defined MET signaling pathway. Our initial decision to examine the RTK MET gene as an ASD risk candidate was based on several factors, including the location of the gene under a broad linkage peak on chromosome 7 that has been replicated multiple times (68–72) as a region carrying ASD risk genes, as well as a number of developmental neurobiology findings that implicate MET signaling in forebrain circuit development. MET activation by HGF modulates forebrain interneuron motility in vitro (73). Excitatory/inhibitory imbalance has been postulated to occur in ASD (74, 75). Moreover, in mice gene targeting of the MET signaling pathway, through deletion of the gene encoding plasminogen activator, urokinase receptor (Plaur), which controls levels of HGF, results in reduced numbers of neocortical interneurons, spontaneous seizures (which occur in 20%–30% of children with ASD), increased anxiety, and reduced social interactions (76–78). Additionally, MET signaling participates in autonomic nervous system and cerebellar development, immune function, and gastrointestinal function and repair (79–84). Disruptions of these neural and peripheral elements have been reported in ASD (85–90).
Candidate gene analyses often fail to generate replicable findings due to small effects in a limited number of samples. In the case of the MET signaling cascade (Figure 2), however, the pathophysiologic and genetic evidence of its contribution to ASD risk is now considerable. First, the expression of MET protein is reduced by approximately 2-fold in the postmortem temporal neocortex of individuals with ASD compared with age- and gender-matched controls (52). Second, several thousand samples have been genotyped to reveal significant association of the MET promoter rs1858830 C allele with ASD risk in a 204-family Italian cohort, a larger 539-family US replication cohort (26), and a third, distinct cohort of 101 US families (27). Third, the ASD-associated MET promoter allele is functional in cell-based assays, reducing dramatically the binding of the transcription factor SP1 to the MET promoter and reducing transcription from the MET promoter by approximately 2-fold (26). Fourth, the rs38845 A allele in intron 1 of MET is associated with ASD risk in a cohort of 335 International Molecular Genetic Study of Autism Consortium families (91). The same research group replicated association of this allele with ASD risk in an Italian case-control sample (91). Fifth, in addition to the SNP alleles that may regulate MET transcription, 2 of 26 cases with ASD in which rare de novo CNV losses were observed, had CNV losses of the chromosome 7 region, including the MET gene (25). Sixth, although not statistically significant, direct resequencing of the 21 MET exons in several hundred cases and controls identified functional mutations that are more prevalent in the cases compared with controls (26). The mutations alter the juxtamembrane region of MET, which regulates receptor activation (92). Last, five other genes in the MET signaling pathway were examined in a large family cohort. Two of the genes, PLAUR and serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 (SERPINE1), are associated with increased ASD risk (27), and each mRNA exhibits altered expression in the postmortem cerebral cortex of individuals with ASD compared with age- and gender-matched controls (52).
The genetic findings from our own studies and those gathered from analysis of defined neurodevelopmental syndromic disorders implicate PI3K signal disruption in both multigenic and syndromic ASD. The observation that de novo CNV is substantially more common in simplex families than multiplex families (24, 25) suggests that private mutations, along with other rare functional mutations, may contribute to ASD. In contrast, association of the risk alleles in MET (26, 91) and the MET-regulating genes PLAUR and SERPINE1 (27) is found only in multiplex families and is therefore linked to multigenic, idiopathic ASD. Heritability of common risk alleles in multiplex families also is reflected in behavioral profiles in parents of children with ASD. Thus, the broader autism phenotype is found to a far greater extent in parents from multiplex compared with simplex families (28, 29), suggesting that heritable, rather than de novo mechanisms for ASD expression occur in multiplex families. Collectively, there seem to be a number of different genetic etiologies that can contribute to altering MET signaling in many individuals with ASD.
-
G X E interactions in ASD etiology
Beyond multiple genetic elements implicated in ASD risk, the MET/PI3K pathway also is highly vulnerable to environmental perturbations. For example, increasing the redox state of oligodendrocyte progenitor cells by brief exposure to lead or mercury activates c-Cbl–regulated internalization and degradation of certain RTKs, including MET, EGFR, and PDGFR (93). Not all RTKs are affected by stressing cells through altering redox state. The reason for selective vulnerability of MET and other RTKs is not known. Irrespective of the mechanism, the cell stressor results in reduced signaling through ERK/PI3K. Benzo(a)pyrine (BaP), a common chemical in vehicle exhaust, paper and wood processing, and trash incineration, disrupts the binding of transcription factors such as SP1 to DNA targets (94, 95). This is relevant to MET expression and perhaps ASD, because the normal level of binding of SP1 is reduced by the ASD-associated rs1858830 C allele (25). A testable hypothesis would be to combine the MET risk allele with exposure to BaP to examine how the double hit affects expression levels. The findings from the studies involving cell stressors and toxic chemicals suggest additional ways in which genetic risk due to regulatory alleles may combine with environmental factors to shift a system closer to disease threshold (Figure 3).
-
Final thoughts
ASD heterogeneity needs to be considered more seriously in developing strategies to investigate underlying biological etiologies. Within syndromic and multigenic ASDs, functional profiles are diverse. Heterogeneity at the genetic level may be probed more strategically by using much larger sample populations, as has been done for diabetes and cardiovascular disease. Technology development will continue to facilitate the larger scale association and deep sequencing studies that will generate new candidates and validate current risk factors. We believe it is important not to lose sight of the long-term challenge, which will be to translate the genetic risk of ASD into biologically plausible mechanisms (Figure 1) that can lead to earlier diagnosis and individualized treatments. Although a number of signaling pathways are likely to be involved, the recent surge of convergent findings on the MET and ERK/PI3K signaling pathways, together with the data implicating key structural and adhesion proteins in synapse formation and maturation brings us closer to defining one cellular process that may serve to help us identify new, credible biomarkers and treatment targets.
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: ASD, autism spectrum disorder; CNV, copy number variation; G X E, gene by environment; MET, met proto-oncogene; mTOR, mammalian target of rapamycin; RTK, receptor tyrosine kinase.
Reference information: J. Clin. Invest.119:747–754 (2009). doi:10.1172/JCI37934
Pat Levitt’s present address is: Zilkha Neurogenetic Institute, Keck School of Medicine of University of Southern California, Los Angeles, California, USA.
-
References
- Newschaffer, C.J., et al. 2007. The epidemiology of autism spectrum disorders. Annu. Rev. Public Health. 28:235-258.
- American Psychiatric Association. 1994.Diagnostic and statistical manual of mental disorders. 4th edition. American Psychiatric Association. Washington, DC, USA. 886 pp.
- Knudsen, E.I. 2004. Sensitive periods in the development of the brain and behavior. J. Cogn. Neurosci. 16:1412-1425.
- Thatcher, R. 1994. Psychopathology of early frontal lobe damage: dependence on cycles of
development. Dev. Psychopathol. 6:565-596. View this article via: CrossRef Google Scholar
- Hammock, E.A.D., Levitt, P. 2006. The discipline of neurobehavioral development: the emerging interface of processes that build circuits and skills. Hum. Dev. 49:294-309.
- Dawson, G., et al. 2002. Defining the broader phenotype of autism: genetic, brain, and
behavioral perspectives. Dev. Psychopathol. 14:581-611. View this article via: PubMed Google Scholar
- Beauchaine, T.P., Strassberg, Z., Kees, M.R., Drabick, D.A. 2002. Cognitive response repertoires to child noncompliance by mothers of aggressive boys. J. Abnorm. Child Psychol. 30:89-101.
- Piven, J. 2001. The broad autism phenotype: a complementary strategy for molecular genetic studies of autism. Am. J. Med. Genet. 105:34-35.
- Devlin, B., et al. 2005. Autism and the serotonin transporter: the long and short of it. Mol. Psychiatry. 10:1110-1116.
- Rutter, M. 1996. Autism research: prospects and priorities. J. Autism Dev. Disord. 26:257-275.
- Volkmar, F.R., Lord, C., Bailey, A., Schultz, R.T., Klin, A. 2004. Autism and pervasive developmental disorders. J. Child Psychol. Psychiatry. 45:135-170.
- Bailey, A., et al. 1995. Autism as a strongly genetic disorder: evidence from a British twin
study. Psychol. Med. 25:63-77. View this article via: PubMed Google Scholar
- Steffenburg, S., et al. 1989. A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J. Child Psychol. Psychiatry. 30:405-416.
- Geschwind, D.H., Levitt, P. 2007. Autism spectrum disorders: developmental disconnection syndromes. Curr. Opin. Neurobiol. 17:103-111.
- Veenstra-Vanderweele, J., Christian, S.L., Cook (Jr.), E.H. 2004. Autism as a paradigmatic complex genetic disorder. Annu. Rev. Genomics Hum. Genet. 5:379-405.
- Abrahams, B.S., Geschwind, D.H. 2008. Advances in autism genetics: on the threshold of a new neurobiology. Nat. Rev. Genet. 9:341-355.
- O’Roak, B.J., State, M.W. 2008. Autism genetics: strategies, challenges, and opportunities. Autism Res. 1:4-17. View this article via: CrossRef Google Scholar
- Craddock, N., O’Donovan, M.C., Owen, M.J. 2008. Genome-wide association studies in psychiatry: lessons from early studies of non-psychiatric and psychiatric phenotypes. Mol. Psychiatry. 13:649-653.
- Cook (Jr.), E.H., Scherer, S.W. 2008. Copy-number variations associated with neuropsychiatric conditions. Nature. 455:919-923.
- Sebat, J., et al. 2004. Large-scale copy number polymorphism in the human genome. Science. 305:525-528.
- Rehen, S.K., et al. 2005. Constitutional aneuploidy in the normal human brain. J. Neurosci. 25:2176-2180.
- Kingsbury, M.A., Yung, Y.C., Peterson, S.E., Westra, J.W., Chun, J. 2006. Aneuploidy in the normal and diseased brain. Cell. Mol. Life Sci. 63:2626-2641.
- Kingsbury, M.A., et al. 2005. Aneuploid neurons are functionally active and integrated into brain circuitry. Proc. Natl. Acad. Sci. U. S. A. 102:6143-6147.
- Sebat, J., et al. 2007. Strong association of de novo copy number mutations with autism. Science. 316:445-449.
- Marshall, C.R., et al. 2008. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 82:477-488.
- Campbell, D.B., et al. 2006. A genetic variant that disrupts MET transcription is associated with autism. Proc. Natl. Acad. Sci. U. S. A. 103:16834-16839.
- Campbell, D.B., Li, C., Sutcliffe, J.S., Persico, A.M., Levitt, P. 2008. Genetic evidence implicating multiple genes in the MET receptor
tyrosine kinase pathway in autism spectrum disorder. Autism Res. 1:159-168. View this article via: CrossRef Google Scholar
- Losh, M., Sullivan, P.F., Trembath, D., Piven, J. 2008. Current developments in the genetics of autism: from phenome to genome. J. Neuropathol. Exp. Neurol. 67:829-837.
- Liu, X.Q., Paterson, A.D., Szatmari, P. 2008. Genome-wide linkage analyses of quantitative and categorical autism subphenotypes. Biol. Psychiatry. 64:561-570.
- Daiger, S.P., Bowne, S.J., Sullivan, L.S. 2007. Perspective on genes and mutations causing retinitis pigmentosa. Arch. Ophthalmol. 125:151-158.
- Porter, F.D. 2008. Smith-Lemli-Opitz syndrome: pathogenesis, diagnosis and management. Eur. J. Hum. Genet. 16:535-541.
- Todd, J.A., et al. 2007. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 39:857-864.
- Zeggini, E., et al. 2007. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 316:1336-1341.
- Samani, N.J., et al. 2007. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 357:443-453.
- Bakkaloglu, B., et al. 2008. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am. J. Hum. Genet. 82:165-173.
- Kates, W.R., et al. 2007. Comparing phenotypes in patients with idiopathic autism to patients with velocardiofacial syndrome (22q11 DS) with and without autism. Am. J. Med. Genet. A. 143A:2642-2650.
- Gothelf, D., et al. 2008. Neuroanatomy of fragile X syndrome is associated with aberrant behavior and the fragile X mental retardation protein (FMRP). Ann. Neurol. 63:40-51.
- Nishimura, Y., et al. 2007. Genome-wide expression profiling of lymphoblastoid cell lines distinguishes different forms of autism and reveals shared pathways. Hum. Mol. Genet. 16:1682-1698.
- Persico, A., Bourgeron, T. 2006. Searching for ways out of the autism maze: genetic, epigenetic and environmental clues. Trends Neurosci. 29:349-358.
- Walsh, C.A., Morrow, E.M., Rubenstein, J.L.R. 2008. Autism and brain development. Cell. 135:396-400.
- Zoghbi, H.Y. 2003. Postnatal neurodevelopmental disorders: meeting at the synapse? Science. 302:826-830.
- Urdinguio, R.G., et al. 2008. Mecp2-null mice provide new neuronal targets for Rett syndrome. PLoS ONE. 3:e3669. View this article via: PubMed Google Scholar
- Kwon, C.H., Zhu, X., Zhang, J., Baker, S.J. 2003. mTor is required for hypertrophy of Pten-deficient neuronal soma in vivo. Proc. Natl. Acad. Sci. U. S. A. 100:12923-12928.
- Meikle, L., et al. 2008. Response of a neuronal model of tuberous sclerosis to mammalian target of rapamycin (mTOR) inhibitors: effects on mTORC1 and Akt signaling lead to improved survival and function. J. Neurosci. 28:5422-5432.
- Ehninger, D., et al. 2008. Reversal of learning deficits in a Tsc2+/– mouse model of tuberous sclerosis. Nat. Med. 14:843-848.
- Costa, R.M., et al. 2002. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature. 415:526-530.
- Li, W., et al. 2005. The HMG-CoA reductase inhibitor lovastatin reverses the learning and attention deficits in a mouse model of neurofibromatosis type 1. Curr. Biol. 15:1961-1967.
- Network, T.C.G.A.R. 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 455:1061-1068.
- Ding, L., et al. 2008. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 455:1069-1075.
- Chalhoub, N., Baker, S.J. 2008. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol.Online publication ahead of print. doi:
- Serajee, F.J., Nabi, R., Zhong, H., Mahbubul Huq, A.H. 2003. Association of INPP1, PIK3CG, and TSC2 gene variants with autistic
disorder: implications for phosphatidylinositol signalling in autism. J. Med. Genet. 40:e119. View this article via: PubMed Google Scholar
- Campbell, D.B., et al. 2007. Disruption of cerebral cortex MET signaling in autism spectrum disorder. Ann. Neurol. 62:243-250.
- Harrison, P.J., Law, A.J. 2006. Neuregulin 1 and schizophrenia: genetics, gene expression, and neurobiology. Biol. Psychiatry. 60:132-140.
- Kanakry, C.G., Li, Z., Nakai, Y., Sei, Y., Weinberger, D.R. 2007. Neuregulin-1 regulates cell adhesion via an ErbB2/phosphoinositide-3
kinase/Akt-dependent pathway: potential implications for schizophrenia and cancer. PLoS ONE. 2:e1369. View this article via: PubMed Google Scholar
- Grinshpoon, A., et al. 2005. Cancer in schizophrenia: is the risk higher or lower? Schizophr. Res. 73:333-341.
- Hippisley-Cox, J., Vinogradova, Y., Coupland, C., Parker, C. 2007. Risk of malignancy in patients with schizophrenia or bipolar disorder: nested case-control study. Arch. Gen. Psychiatry. 64:1368-1376.
- Lichtermann, D., Ekelund, J., Pukkala, E., Tanskanen, A., Lonnqvist, J. 2001. Incidence of cancer among persons with schizophrenia and their relatives. Arch. Gen. Psychiatry. 58:573-578.
- Levav, I., et al. 2007. Cancer risk among parents and siblings of patients with schizophrenia. Br. J. Psychiatry. 190:156-161.
- Catts, V.S., Catts, S.V., O’Toole, B.I., Frost, A.D. 2008. Cancer incidence in patients with schizophrenia and their first-degree relatives - a meta-analysis. Acta Psychiatr. Scand. 117:323-336.
- Norton, N., et al. 2006. Evidence that interaction between neuregulin 1 and its receptor erbB4 increases susceptibility to schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 141B:96-101.
- Fox, I.J., Kornblum, H.I. 2005. Developmental profile of ErbB receptors in murine central nervous system: implications for functional interactions. J. Neurosci. Res. 79:584-597.
- Judson, M.C., Bergman, M.Y., Campbell, D.B., Eagleson, K.L., Levitt, P. 2009. Dynamic gene and protein expression patterns of the autism-associated c-Met receptor tyrosine kinase in the developing mouse forebrain. J. Comp. Neurol. 513:511-531.
- Hossain, M.A., Russell, J.C., Gomez, R., Laterra, J. 2002. Neuroprotection by scatter factor/hepatocyte growth factor and FGF-1 in cerebellar granule neurons is phosphatidylinositol 3-kinase/akt-dependent and MAPK/CREB-independent. J. Neurochem. 81:365-378.
- Liu, Y., et al. 2007. Hepatocyte growth factor and c-Met expression in pericytes: implications for atherosclerotic plaque development. J. Pathol. 212:12-19.
- Roggia, C., Ukena, C., Bohm, M., Kilter, H. 2007. Hepatocyte growth factor (HGF) enhances cardiac commitment of differentiating embryonic stem cells by activating PI3 kinase. Exp. Cell Res. 313:921-930.
- Cacci, E., et al. 2003. Hepatocyte growth factor stimulates cell motility in cultures of the striatal progenitor cells ST14A. J. Neurosci. Res. 74:760-768.
- He, F., et al. 2008. HGF protects cultured cortical neurons against hypoxia/reoxygenation induced cell injury via ERK1/2 and PI-3K/Akt pathways. Colloids Surf. B Biointerfaces. 61:290-297.
- [No authors listed]. 1998. A full genome screen for autism with evidence for linkage to a region on chromosome 7q. International Molecular Genetic Study of Autism Consortium. Hum. Mol. Genet. 7:571-578.
- Philippe, A., et al. 1999. Genome-wide scan for autism susceptibility genes. Paris Autism Research International Sibpair Study. Hum. Mol. Genet. 8:805-812.
- International Molecular Genetic Study of Autism Consortium. 2001. A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am. J. Hum. Genet. 69:570-581.
- Lamb, J.A., et al. 2005. Analysis of IMGSAC autism susceptibility loci: evidence for sex limited and parent of origin specific effects. J. Med. Genet. 42:132-137.
- Schellenberg, G.D., et al. 2006. Evidence for multiple loci from a genome scan of autism kindreds. Mol. Psychiatry. 11:1049-1060.
- Powell, E.M., Mars, W.M., Levitt, P. 2001. Hepatocyte growth factor/scatter factor is a motogen for interneurons migrating from the ventral to dorsal telencephalon. Neuron. 30:79-89.
- Levitt, P., Eagleson, K.L., Powell, E.M. 2004. Regulation of neocortical interneuron development and the implications for neurodevelopmental disorders. Trends Neurosci. 27:400-406.
- Rubenstein, J.L., Merzenich, M.M. 2003. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2:255-267.
- Powell, E.M., et al. 2003. Genetic disruption of cortical interneuron development causes region-
and GABA cell type-specific deficits, epilepsy, and behavioral dysfunction. J. Neurosci. 23:622-631. View this article via: PubMed Google Scholar
- Levitt, P. 2005. Disruption of interneuron development. Epilepsia. 46(Suppl. 7):22-28. View this article via: PubMed Google Scholar
- Eagleson, K.L., Bonnin, A., Levitt, P. 2005. Region- and age-specific deficits in gamma-aminobutyric acidergic neuron development in the telencephalon of the uPAR(–/–) mouse. J. Comp. Neurol. 489:449-466.
- Beilmann, M., et al. 1997. Neoexpression of the c-met/hepatocyte growth factor-scatter factor
receptor gene in activated monocytes. Blood. 90:4450-4458. View this article via: PubMed Google Scholar
- Ieraci, A., Forni, P.E., Ponzetto, C. 2002. Viable hypomorphic signaling mutant of the Met receptor reveals a role for hepatocyte growth factor in postnatal cerebellar development. Proc. Natl. Acad. Sci. U. S. A. 99:15200-15205.
- Arthur, L.G., Schwartz, M.Z., Kuenzler, K.A., Birbe, R. 2004. Hepatocyte growth factor treatment ameliorates diarrhea and bowel inflammation in a rat model of inflammatory bowel disease. J. Pediatr. Surg. 39:139-143.
- Okunishi, K., et al. 2005. A novel role of hepatocyte growth factor as an immune regulator
through suppressing dendritic cell function. J. Immunol. 175:4745-4753. View this article via: PubMed Google Scholar
- Ido, A., Numata, M., Kodama, M., Tsubouchi, H. 2005. Mucosal repair and growth factors: recombinant human hepatocyte growth factor as an innovative therapy for inflammatory bowel disease. J. Gastroenterol. 40:925-931.
- McCall-Culbreath, K.D., Li, Z., Zutter, M.M. 2008. Crosstalk between the alpha2beta1 integrin and c-met/HGF-R regulates innate immunity. Blood. 111:3562-3570.
- Jyonouchi, H., Geng, L., Ruby, A., Zimmerman-Bier, B. 2005. Dysregulated innate immune responses in young children with autism spectrum disorders: their relationship to gastrointestinal symptoms and dietary intervention. Neuropsychobiology. 51:77-85.
- Valicenti-McDermott, M., et al. 2006. Frequency of gastrointestinal symptoms in children with autistic spectrum disorders and association with family history of autoimmune disease. J. Dev. Behav. Pediatr. 27:S128-S136.
- Hansen, R.L., et al. 2008. Regression in autism: prevalence and associated factors in the CHARGE Study. Ambul. Pediatr. 8:25-31.
- Xue, M., Brimacombe, M., Chaaban, J., Zimmerman-Bier, B., Wagner, G.C. 2008. Autism spectrum disorders: concurrent clinical disorders. J. Child Neurol. 23:6-13.
- Garbett, K., et al. 2008. Immune transcriptome alterations in the temporal cortex of subjects with autism. Neurobiol. Dis. 30:303-311.
- Enstrom, A.M., et al. 2009. Altered gene expression and function of peripheral blood natural killer cells in children with autism. Brain Behav. Immun. 23:124-133.
- Sousa, I., et al. 2008. MET and autism susceptibility: family and case-control studies. Eur. J. Hum. Genet. Online publication ahead of print. doi: 10.1038/ejhg.2008.215.
- Ma, P.C., et al. 2003. c-MET mutational analysis in small cell lung cancer: novel
juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res. 63:6272-6281. View this article via: PubMed Google Scholar
- Li, Z., Dong, T., Proschel, C., Noble, M. 2007. Chemically diverse toxicants converge on Fyn and c-Cbl to disrupt
precursor cell function. PLoS Biol. 5:e35. View this article via: PubMed Google Scholar
- Hood, D.B., Nayyar, T., Ramesh, A., Greenwood, M., Inyang, F. 2000. Modulation in the developmental expression profile of Sp1 subsequent to transplacental exposure of fetal rats to desorbed benzo[a]pyrene following maternal inhalation. Inhal. Toxicol. 12:511-535.
- Nayyar, T., Zawia, N.H., Hood, D.B. 2002. Transplacental effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin on the temporal modulation of Sp1 DNA binding in the developing cerebral cortex and cerebellum. Exp. Toxicol. Pathol. 53:461-468.
- Peters, S.U., Beaudet, A.L., Madduri, N., Bacino, C.A. 2004. Autism in Angelman syndrome: implications for autism research. Clin. Genet. 66:530-536.
- Veltman, M.W., Craig, E.E., Bolton, P.F. 2005. Autism spectrum disorders in Prader-Willi and Angelman syndromes: a systematic review. Psychiatr. Genet. 15:243-254.
- Bonati, M.T., et al. 2007. Evaluation of autism traits in Angelman syndrome: a resource to unfold autism genes. Neurogenetics. 8:169-178.
- Lowenthal, R., Paula, C.S., Schwartzman, J.S., Brunoni, D., Mercadante, M.T. 2007. Prevalence of pervasive developmental disorder in Down’s syndrome. J. Autism Dev. Disord. 37:1394-1395.
- Kent, L., Evans, J., Paul, M., Sharp, M. 1999. Comorbidity of autistic spectrum disorders in children with Down syndrome. Dev. Med. Child Neurol. 41:153-158.
- Garcia-Nonell, C., et al. 2008. Secondary medical diagnosis in fragile X syndrome with and without autism spectrum disorder. Am. J. Med. Genet. A. 146A:1911-1916.
- Clifford, S., et al. 2007. Autism spectrum phenotype in males and females with fragile X full mutation and premutation. J. Autism Dev. Disord. 37:738-747.
- Williams, P.G., Hersh, J.H. 1998. Brief report: the association of neurofibromatosis type 1 and autism. J. Autism Dev. Disord. 28:567-571.
- Butler, M.G., et al. 2005. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J. Med. Genet. 42:318-321.
- Potocki, L., et al. 2007. Characterization of Potocki-Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am. J. Hum. Genet. 80:633-649.
- Chahrour, M., Zoghbi, H.Y. 2007. The story of Rett syndrome: from clinic to neurobiology. Neuron. 56:422-437.
- Ben Zeev Ghidoni, B. 2007. Rett syndrome. Child Adolesc. Psychiatr. Clin. N. Am. 16:723-743.
- Tierney, E., et al. 2001. Behavior phenotype in the RSH/Smith-Lemli-Opitz syndrome. Am J. Med. Genet. 98:191-200.
- Sikora, D.M., Pettit-Kekel, K., Penfield, J., Merkens, L.S., Steiner, R.D. 2006. The near universal presence of autism spectrum disorders in children
with Smith-Lemli-Opitz syndrome. Am J. Med. Genet. A. 140:1511-1518. View this article via: PubMed Google Scholar
- Splawski, I., et al. 2004. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 119:19-31.
- Smalley, S.L. 1998. Autism and tuberous sclerosis. J. Autism Dev. Disord. 28:407-414.
- Jeste, S.S., Sahin, M., Bolton, P., Ploubidis, G.B., Humphrey, A. 2008. Characterization of autism in young children with tuberous sclerosis complex. J. Child Neurol. 23:520-525.
- Philippe, A., et al. 2008. Neurobehavioral profile and brain imaging study of the 22q13.3
deletion syndrome in childhood. Pediatrics. 122:e376-e382. View this article via: PubMed Google Scholar
-
Version history
- Version 1 (April 1, 2009): No description
Article tools
-
View PDF

- Download citation information
- Send a comment
- Terms of use
- Standard abbreviations
- Need help? Email the journal
Review Series
Rethinking Mental Disorders
-
Schizophrenia from a neural circuitry perspective: advancing toward rational pharmacological therapies
David A. Lewis et al. -
Children with obsessive-compulsive disorder: are they just “little adults”?
Simran K. Kalra et al. -
Bipolar disorder: from genes to behavior pathways
Keri Martinowich et al. -
The genetic and neurobiologic compass points toward common signaling dysfunctions in autism spectrum disorders
Pat Levitt et al. -
Targeted electrode-based modulation of neural circuits for depression
Helen S. Mayberg -
Disruptive insights in psychiatry: transforming a clinical discipline
Thomas R. Insel
Metrics
Go to
- Top
- Abstract
- Distinct genetic mechanisms can result in ASD
- Distinct patterns of heritability of risk alleles in ASD
- Syndromic disorders and rare mutations point the way
- Intracellular kinase signaling in ASD-associated disorders
- MET in PI3K signaling and ASD
- G X E interactions in ASD etiology
- Final thoughts
- Footnotes
- References
- Version history



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)




