Advertisement
Research ArticleCell biologyInfectious disease Free access | 10.1172/JCI129679
sEH promotes macrophage phagocytosis and lung clearance of Streptococcus pneumoniae
Hong Li, J. Alyce Bradbury, Matthew L. Edin, Joan P. Graves, Artiom Gruzdev, Jennifer Cheng, Samantha L. Hoopes, Laura M. DeGraff, Michael B. Fessler, Stavros Garantziotis, Shepherd H. Schurman, and Darryl C. Zeldin
Division of Intramural Research, National Institute of Environmental Health Sciences (NIEHS), NIH, Research Triangle Park, North Carolina, USA.
Address correspondence to: Darryl C. Zeldin, Division of Intramural Research, NIH/NIEHS, 111 T.W. Alexander Drive, Building 101, Room A214, Research Triangle Park, North Carolina 27709, USA. Phone: 984.287.3641; Email: zeldin@niehs.nih.gov.
Authorship note: HL, AB, and MLE are co–first authors and contributed equally to this work.
Find articles by Li, H. in: JCI | PubMed | Google Scholar
Division of Intramural Research, National Institute of Environmental Health Sciences (NIEHS), NIH, Research Triangle Park, North Carolina, USA.
Address correspondence to: Darryl C. Zeldin, Division of Intramural Research, NIH/NIEHS, 111 T.W. Alexander Drive, Building 101, Room A214, Research Triangle Park, North Carolina 27709, USA. Phone: 984.287.3641; Email: zeldin@niehs.nih.gov.
Authorship note: HL, AB, and MLE are co–first authors and contributed equally to this work.
Find articles by Bradbury, J. in: JCI | PubMed | Google Scholar
Division of Intramural Research, National Institute of Environmental Health Sciences (NIEHS), NIH, Research Triangle Park, North Carolina, USA.
Address correspondence to: Darryl C. Zeldin, Division of Intramural Research, NIH/NIEHS, 111 T.W. Alexander Drive, Building 101, Room A214, Research Triangle Park, North Carolina 27709, USA. Phone: 984.287.3641; Email: zeldin@niehs.nih.gov.
Authorship note: HL, AB, and MLE are co–first authors and contributed equally to this work.
Find articles by
Edin, M.
in:
JCI
|
PubMed
|
Google Scholar
|

Division of Intramural Research, National Institute of Environmental Health Sciences (NIEHS), NIH, Research Triangle Park, North Carolina, USA.
Address correspondence to: Darryl C. Zeldin, Division of Intramural Research, NIH/NIEHS, 111 T.W. Alexander Drive, Building 101, Room A214, Research Triangle Park, North Carolina 27709, USA. Phone: 984.287.3641; Email: zeldin@niehs.nih.gov.
Authorship note: HL, AB, and MLE are co–first authors and contributed equally to this work.
Find articles by Graves, J. in: JCI | PubMed | Google Scholar
Division of Intramural Research, National Institute of Environmental Health Sciences (NIEHS), NIH, Research Triangle Park, North Carolina, USA.
Address correspondence to: Darryl C. Zeldin, Division of Intramural Research, NIH/NIEHS, 111 T.W. Alexander Drive, Building 101, Room A214, Research Triangle Park, North Carolina 27709, USA. Phone: 984.287.3641; Email: zeldin@niehs.nih.gov.
Authorship note: HL, AB, and MLE are co–first authors and contributed equally to this work.
Find articles by
Gruzdev, A.
in:
JCI
|
PubMed
|
Google Scholar
|

Division of Intramural Research, National Institute of Environmental Health Sciences (NIEHS), NIH, Research Triangle Park, North Carolina, USA.
Address correspondence to: Darryl C. Zeldin, Division of Intramural Research, NIH/NIEHS, 111 T.W. Alexander Drive, Building 101, Room A214, Research Triangle Park, North Carolina 27709, USA. Phone: 984.287.3641; Email: zeldin@niehs.nih.gov.
Authorship note: HL, AB, and MLE are co–first authors and contributed equally to this work.
Find articles by
Cheng, J.
in:
JCI
|
PubMed
|
Google Scholar
|

Division of Intramural Research, National Institute of Environmental Health Sciences (NIEHS), NIH, Research Triangle Park, North Carolina, USA.
Address correspondence to: Darryl C. Zeldin, Division of Intramural Research, NIH/NIEHS, 111 T.W. Alexander Drive, Building 101, Room A214, Research Triangle Park, North Carolina 27709, USA. Phone: 984.287.3641; Email: zeldin@niehs.nih.gov.
Authorship note: HL, AB, and MLE are co–first authors and contributed equally to this work.
Find articles by Hoopes, S. in: JCI | PubMed | Google Scholar
Division of Intramural Research, National Institute of Environmental Health Sciences (NIEHS), NIH, Research Triangle Park, North Carolina, USA.
Address correspondence to: Darryl C. Zeldin, Division of Intramural Research, NIH/NIEHS, 111 T.W. Alexander Drive, Building 101, Room A214, Research Triangle Park, North Carolina 27709, USA. Phone: 984.287.3641; Email: zeldin@niehs.nih.gov.
Authorship note: HL, AB, and MLE are co–first authors and contributed equally to this work.
Find articles by DeGraff, L. in: JCI | PubMed | Google Scholar
Division of Intramural Research, National Institute of Environmental Health Sciences (NIEHS), NIH, Research Triangle Park, North Carolina, USA.
Address correspondence to: Darryl C. Zeldin, Division of Intramural Research, NIH/NIEHS, 111 T.W. Alexander Drive, Building 101, Room A214, Research Triangle Park, North Carolina 27709, USA. Phone: 984.287.3641; Email: zeldin@niehs.nih.gov.
Authorship note: HL, AB, and MLE are co–first authors and contributed equally to this work.
Find articles by
Fessler, M.
in:
JCI
|
PubMed
|
Google Scholar
|

Division of Intramural Research, National Institute of Environmental Health Sciences (NIEHS), NIH, Research Triangle Park, North Carolina, USA.
Address correspondence to: Darryl C. Zeldin, Division of Intramural Research, NIH/NIEHS, 111 T.W. Alexander Drive, Building 101, Room A214, Research Triangle Park, North Carolina 27709, USA. Phone: 984.287.3641; Email: zeldin@niehs.nih.gov.
Authorship note: HL, AB, and MLE are co–first authors and contributed equally to this work.
Find articles by
Garantziotis, S.
in:
JCI
|
PubMed
|
Google Scholar
|

Division of Intramural Research, National Institute of Environmental Health Sciences (NIEHS), NIH, Research Triangle Park, North Carolina, USA.
Address correspondence to: Darryl C. Zeldin, Division of Intramural Research, NIH/NIEHS, 111 T.W. Alexander Drive, Building 101, Room A214, Research Triangle Park, North Carolina 27709, USA. Phone: 984.287.3641; Email: zeldin@niehs.nih.gov.
Authorship note: HL, AB, and MLE are co–first authors and contributed equally to this work.
Find articles by
Schurman, S.
in:
JCI
|
PubMed
|
Google Scholar
|

Division of Intramural Research, National Institute of Environmental Health Sciences (NIEHS), NIH, Research Triangle Park, North Carolina, USA.
Address correspondence to: Darryl C. Zeldin, Division of Intramural Research, NIH/NIEHS, 111 T.W. Alexander Drive, Building 101, Room A214, Research Triangle Park, North Carolina 27709, USA. Phone: 984.287.3641; Email: zeldin@niehs.nih.gov.
Authorship note: HL, AB, and MLE are co–first authors and contributed equally to this work.
Find articles by Zeldin, D. in: JCI | PubMed | Google Scholar
Authorship note: HL, AB, and MLE are co–first authors and contributed equally to this work.
Published September 30, 2021 - More info
J Clin Invest. 2021;131(22):e129679. https://doi.org/10.1172/JCI129679.
© 2021 American Society for Clinical Investigation
Received: May 1, 2019; Accepted: September 28, 2021
-
Abstract
Epoxyeicosatrienoic acids (EETs) have potent antiinflammatory properties. Hydrolysis of EETs by soluble epoxide hydrolase/ epoxide hydrolase 2 (sEH/EPHX2) to less active diols attenuates their antiinflammatory effects. Macrophage activation is critical to many inflammatory responses; however, the role of EETs and sEH in regulating macrophage function remains unknown. Lung bacterial clearance of Streptococcus pneumoniae was impaired in Ephx2-deficient (Ephx2–/–) mice and in mice treated with an sEH inhibitor. The EET receptor antagonist EEZE restored lung clearance of S. pneumoniae in Ephx2–/– mice. Ephx2–/– mice had normal lung Il1b, Il6, and Tnfa expression levels and macrophage recruitment to the lungs during S. pneumoniae infection; however, Ephx2 disruption attenuated proinflammatory cytokine induction, Tlr2 and Pgylrp1 receptor upregulation, and Ras-related C3 botulinum toxin substrates 1 and 2 (Rac1/2) and cell division control protein 42 homolog (Cdc42) activation in PGN-stimulated macrophages. Consistent with these observations, Ephx2–/– macrophages displayed reduced phagocytosis of S. pneumoniae in vivo and in vitro. Heterologous overexpression of TLR2 and peptidoglycan recognition protein 1 (PGLYRP1) in Ephx2–/– macrophages restored macrophage activation and phagocytosis. Human macrophage function was similarly regulated by EETs. Together, these results demonstrate that EETs reduced macrophage activation and phagocytosis of S. pneumoniae through the downregulation of TLR2 and PGLYRP1 expression. Defining the role of EETs and sEH in macrophage function may lead to the development of new therapeutic approaches for bacterial diseases.
-
Introduction
Pneumonia is the leading cause of mortality in the United States. Streptococcus pneumoniae and Klebsiella pneumoniae are the most frequently isolated bacterial pathogens in community-acquired pneumonia (1). Together, these account for more deaths (nearly 1.6 million per year) than any other pathogen (2, 3). Therapeutic options are limited by emerging antimicrobial resistance (4). A better understanding of immune defenses against S. pneumoniae and K. pneumoniae is critical to the discovery of novel preventive and therapeutic strategies.
Alveolar macrophages (AMs) are key cells involved in orchestrating the innate immune response to bacterial infection in the lungs (5). These cells patrol gas-exchanging alveoli and remove pathogens through phagocytosis and intracellular killing. Furthermore, AMs alert the host to the presence of invading microbes by releasing lipid and protein mediators that activate other resident cells and recruit neutrophils to the site of infection (6). This response is highly regulated to limit self-inflicted damage to host cells and tissues (7). Emerging evidence suggests that lipid mediators are major regulators of both the amplitude and duration of infection-triggered inflammatory responses (8). For example, leukotriene (LT) B4 has potent proinflammatory effects that augment the innate immune functions of AMs (9), while prostaglandins (PGs) including PGE2 and PGI2 have antiinflammatory actions in the context of bacterial pneumonia (10, 11).
Bacterial clearance during lung infection is mainly accomplished through phagocytosis by alveolar macrophages. Bacterial phagocytosis is initiated through receptors called pattern recognition receptors (PRRs) that recognize pathogen-associated molecular patterns (PAMPs). PAMPs are conserved motifs found in microbial pathogens; examples include LPS in Gram-negative bacteria, peptidoglycan (PGN) and lipoteichoic acid (LTA) in Gram-positive bacteria, and mannan (Man) in yeast. PAMP detection enhances phagocytosis, activates complement cascades, and initiates inflammatory signaling pathways (12). Extracellular pattern recognition molecules including mannose-binding lectin, C-reactive protein, and serum amyloid protein are important in the innate immune response to a variety of microbial infections (13). Cell-surface PRRs such as the TLRs are also essential in innate immune recognition (14). The microbial components recognized by TLRs have been identified for TLR2 (lipoproteins and PGN; ref. 15), TLR3 (double-stranded RNA; ref. 16), TLR4 (LPS; ref. 17), TLR5 (bacterial flagellin; ref. 18), TLR6 (mycoplasmal macrophage-activating lipopeptide-2 kDa; ref. 19), TLR7 and TLR8 (single-stranded RNA; ref. 20), and TLR9 (CpG bacterial DNA; ref. 21). Peptidoglycan recognition protein (PGLYRP) receptors specifically recognize Gram-positive bacteria (22–25). In mammals, there are 4 PGLYRPs: PGLYRP1, PGLYRP2, PGLYRP3, and PGLYRP4. Of these, only PGLYRP1 is significantly expressed in monocytes and macrophages (26). The function and regulation of PGLYRP1 in innate immune responses of macrophages is largely unknown.
Oxygenation of arachidonic acid (AA) by cytochrome P450 (CYP) epoxygenases forms epoxyeicosatrienoic acids (EETs) that exhibit potent antiinflammatory properties (27). EETs are converted to less active dihydroxyeicosatrienoic acids (DHETs) by soluble epoxide hydrolase (sEH, encoded by the Ephx2 gene; ref. 28). sEH is a cytosolic enzyme that catalyzes the hydrolysis of a diverse group of epoxides to their corresponding vicinal diols (29). The broad spectrum of xenobiotic epoxides metabolized by sEH suggests a role in the protection of cells against the potentially harmful effects of these compounds (30). In addition, sEH hydrolyzes endogenous fatty acid epoxides, with AA-derived EETs being among the preferred substrates (31). Therefore, sEH has an important physiological role in regulating steady-state levels of lipid signaling molecules such as EETs.
Genetic disruption of Ephx2 in mice results in increased circulating EET levels; therefore, sEH-deficient (Ephx2–/–) mice are an excellent model with which to examine the function of EETs in vivo (32). Ephx2–/– mice are protected against inflammation induced by bacterial LPS. Compared with lungs from control mice, lungs from Ephx2–/– mice have reduced levels of proinflammatory cytokines, attenuated endothelial cell adhesion molecule expression, and reduced neutrophil infiltration after LPS exposure (33). Pharmacological inhibition of sEH also results in increased EETs, leading to decreased inflammation (34–36). In humans, sEH expression is increased in ulcerative colitis (UC), UC-induced dysplasia, and UC-induced carcinoma (36). Indeed, sEH is a potential biomarker and therapeutic target for inflammation and inflammation-induced carcinoma (36). Although EETs and sEH are known to mediate many inflammatory responses, the role of EETs and sEH in lung bacterial infection is unknown. There is evidence that EET may signal through a putative cell-surface receptor; however, its identity has not been confirmed (37).
In this study, we investigated the regulation of lung bacterial clearance by EETs and sEH. We used Ephx2–/– mice, exogenous administration of EETs or EET antagonists, and transient expression or siRNA suppression of PRRs to determine the molecular mechanisms that regulate bacterial phagocytosis and inflammatory signaling in macrophages. Our data show that Ephx2–/– mice had an impaired innate immune response to the Gram-positive bacteria S. pneumoniae, but not the Gram-positive bacteria Staphylococcus aureus or the Gram-negative bacteria K. pneumoniae. Specifically, EETs suppress induction of the PGN receptors TLR2 and PGLYRP1 to impair inflammatory signaling, which is critical for phagocytosis of S. pneumoniae in vitro and in vivo. We further demonstrate that phagocytosis of S. pneumoniae, proinflammatory cytokine production, and PRR expression in human macrophages were also regulated by EETs and sEH.
-
Results
Ephx2–/– mice have reduced lung bacterial clearance of S. pneumoniae in vivo. Since sEH is highly expressed in lung macrophages (38, 39) and regulates lung inflammation following LPS exposure (33), we examined whether sEH regulates the clearance of bacteria from the lungs. WT and Ephx2–/– mice were infected by intranasal aspiration with either saline (negative control), the Gram-positive bacteria S. pneumoniae, or the Gram-negative bacteria K. pneumoniae, and lung bacterial clearance was determined by serial dilution assay 48 hours later. We observed no significant differences between the WT and Ephx2–/– mice in any of the outcomes measured in saline-treated lungs (Figure 1, A–C). WT and Ephx2–/– mice had similar numbers of K. pneumoniae in their lungs 48 hours after inoculation (Figure 1A). In contrast, lungs from Ephx2–/– mice had significantly increased numbers of S. pneumoniae after 48 hours compared with WT mice, which indicated reduced bacterial clearance (Figure 1A). Consistent with this finding, histological analysis revealed increased inflammation in Ephx2–/– mouse lungs infected with S. pneumoniae relative to WT mouse lungs, with infiltration of neutrophils and monocytes and loss of normal alveolar architecture (Figure 1B). The degree of inflammation was quantified by a pathologist who was blinded to treatment group assignment and genotype. Lungs from Ephx2–/– mice had a significantly higher inflammation score than did lungs from WT mice (2.5 ± 0.8 vs. 1.2 ± 0.5, P < 0.05) after S. pneumoniae infection, but not after K. pneumoniae infection (2.3 ± 0.3 vs. 2.5 ± 0.3, P = 0.53). In the bronchoalveolar lavage fluid (BALF), total cell numbers and macrophage, lymphocyte, and eosinophil numbers were comparable between WT and Ephx2–/– mice infected with S. pneumoniae; however, neutrophil numbers were significantly higher in BALF from infected Ephx2–/– mice compared with BALF from WT mice (Figure 1C).
 Figure 1
Figure 1Impaired bacterial clearance and increased lung inflammation after S. pneumoniae infection in Ephx2–/– mice in vivo. (A) WT and Ephx2–/– mice (n = 6–12 per group) were inoculated by intranasal aspiration with either saline (negative control), K. pneumonia at 2 × 103 CFU/mouse, or S. pneumonia at 2 × 105 CFU/mouse. Colony counts were determined by serial dilution assay in lungs collected 48 hours after inoculation. Each square represents a different animal (white squares represent WT mice; black squares represent Ephx2–/– mice). (B) Representative H&E staining of formalin-fixed lung sections. Scale bars: 200 μm. (C) Bronchoalveolar lavage was performed on the mice in A, and total BALF cells and cell differentials were determined. *P < 0.05, by ordinary 2-way ANOVA, followed by Tukey’s post hoc multiple-comparison test (A and C).
The defect in bacterial clearance in Ephx2–/– mice was selective for S. pneumoniae. WT and Ephx2–/– mice had similar lung bacterial numbers, inflammation score, and proinflammatory cytokine mRNA levels 48 hours after inoculation with a reduced dose of K. pneumoniae (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI129679DS1). In addition, clearance of another Gram-positive bacteria, S. aureus, was not different between WT and Ephx2–/– mice at both high and low inoculation doses, and at both 24 and 48 hours after inoculation (Supplemental Figure 2).
sEH deficiency altered S. pneumoniae clearance as early as 12 hours after infection. Compared with lungs of WT mice, Ephx2–/– mouse lungs had increased numbers of S. pneumoniae after 12 hours (Figure 2A); however, we found no significant differences between WT and Ephx2–/– mice in BALF cells at this time point (Figure 2B). Moreover, Ephx2 disruption did not attenuate S. pneumoniae–induced increases in BALF levels of the proinflammatory cytokines IL-1β, IL-6, or TNF-α (Figure 2C). This suggests that the defect in S. pneumoniae clearance in Ephx2–/– mice was not due to attenuation of pulmonary cytokine induction or suppression of immune cell infiltration into the lungs.
 Figure 2
Figure 2S. pneumoniae lung clearance is impaired early during infection in Ephx2–/– mice and altered by pharmacological sEH inhibition or EET antagonism in vivo. (A) WT and Ephx2–/– mice (n = 5–10 per group) were inoculated by intranasal aspiration with either saline (PBS, negative control) or S. pneumonia at 2 × 105 CFU/mouse. Colony counts were determined by serial dilution assay in lungs collected 12 hours after inoculation. (B) Bronchoalveolar lavage was performed on the mice in A, and total BALF cells and cell differentials were determined. (C) Levels of Il-1β, IL-6, and TNF-α protein in BALF from mice in A were determined by multiplex array. (D) WT mice (n = 14–56 per group) were treated with either vehicle (1% PEG400) or 1% PEG400 containing 10 mg/L TPPU in the drinking water and inoculated by intranasal aspiration with either saline (negative control) or S. pneumonia at 2 × 105 CFU/mouse. Colony counts were determined by serial dilution assay in lungs removed 48 hours after inoculation. Data represent the mean ± SEM of 4 independent experiments. *P < 0.05. (E) WT and Ephx2–/– mice (n = 5–10 per group) were dosed with vehicle or EEZE (15 μg/kg/day) via Alzet minipumps and inoculated with either saline (negative control) or S. pneumonia at 2 × 105 CFU/mouse. Colony counts were determined in lungs removed 48 hours after inoculation. *P < 0.05, by ordinary 2-way ANOVA, followed by Tukey’s post hoc multiple-comparison test (A–E).
We found that clearance of S. pneumoniae was also reduced by treatment with the sEH inhibitor 1-trifluoromethoxyphenyl-3-(1-propionylpiperidin-4-yl) urea (TPPU) before and during infection (Figure 2D). Moreover, treatment of Ephx2–/– mice with the putative EET receptor antagonist 14,15-epoxyeicosa-5(Z)-enoic acid (EEZE) restored the clearance of S. pneumoniae to WT levels (Figure 2E), suggesting that sEH mediated S. pneumoniae clearance through the regulation of epoxy fatty acid hydrolysis. Together, these results indicate that sEH was required for efficient pulmonary clearance of S. pneumoniae, but not K. pneumoniae or S. aureus, and that sEH limited the lung inflammatory response to S. pneumoniae infection.
PAMPs increase sEH expression and EET levels in macrophages in vitro. To determine whether PAMPs alter the expression of sEH, we stimulated isolated peritoneal macrophages from WT mice with various PAMPs and measured sEH expression at the mRNA and protein levels. Macrophage Ephx2 mRNA levels were significantly upregulated by PGN, LTA, and LPS, but not by zymosan (Zym) or Man (Figure 3A). Likewise, sEH protein expression was upregulated in WT macrophages after PGN stimulation (Figure 3B). PGN induced macrophage sEH expression in a dose-dependent manner at concentrations of 0.5–10.0 μg/mL (Figure 3C). Ephx2–/– mice have increased levels of EETs (40), and EETs are known to influence inflammatory responses (41). Thus, sEH may regulate macrophage function during inflammation through metabolism of EETs. To confirm the link between EETs and sEH-mediated macrophage responses to PGN, we initially measured EET levels in WT and Ephx2–/– macrophages by liquid chromatography tandem mass spectrometry (LC-MS/MS). As shown in Figure 3D, PGN significantly increased the levels of EETs in WT macrophages. Importantly, Ephx2–/– macrophages produced higher levels of 8,9-EET, 11,12-EET, and 14,15-EET compared with WT macrophages after PGN treatment. In contrast, the levels of 5,6-EET were not different between the 2 genotypes. These results are consistent with the known regiochemistry of EET metabolism by sEH (39). Together, these data demonstrate that PGN induced sEH expression and increased EET levels in macrophages and that disruption of Ephx2 increased EET levels even further after PGN treatment.
 Figure 3
Figure 3Macrophage sEH expression is regulated by PAMPs in vitro. (A) Ephx2 transcript levels were measured by real-time quantitative RT-PCR in peritoneal macrophages stimulated with vehicle (PBS) or the following PAMPs: LPS (1 μg/mL), LTA (5 μg/mL), PGN (10 μg/mL), Man (10 μg/mL), or Zym (10 μg/mL). n = 5 per group. *P < 0.05 versus vehicle. Immunoblotting for sEH protein expression was done after stimulation of macrophages with different PAMPs (B) or different concentrations of PGN (0–10 μg/mL) (C). Densitometry of sEH protein expression normalized to β-actin was used to quantify the results in B and C. n = 3 per group. *P < 0.05 versus vehicle (none). (D) EET levels were measured by LC-MS/MS in peritoneal macrophages from WT and Ephx2–/– mice with or without 10 μg/mL PGN stimulation. n = 6 per group. *P < 0.05, by ordinary 1-way ANOVA, followed by Tukey’s post hoc multiple-comparison test (A–C) or ordinary 2-way ANOVA, followed by Tukey’s post hoc multiple-comparison test (D).
Ephx2–/– macrophages exhibit impaired bacterial phagocytosis in vitro and in vivo. Since Ephx2–/– mice have normal macrophage numbers and proinflammatory cytokine levels (Figure 1C and Figure 2, B and C) but defective clearance of S. pneumonia (Figure 1A and Figure 2A), we hypothesized that sEH may selectively alter bacterial phagocytosis. To test this hypothesis, we examined phagocytosis of FITC-labeled S. pneumoniae, FITC-labeled K. pneumoniae, and Alexa Fluor 488–labeled S. aureus by peritoneal macrophages from WT and Ephx2–/– mice in vitro. Interestingly, Ephx2–/– macrophages showed defective phagocytosis of S. pneumoniae, but not K. pneumoniae or S. aureus (Figure 4, A–C, Supplemental Figure 1C, and Supplemental Figure 3, A–C). Fluorescence staining confirmed that, compared with WT macrophages, Ephx2–/– macrophages had reduced internalization of S. pneumoniae organisms (Supplemental Figure 4). Ephx2–/– macrophages also showed reduced phagocytosis of S. pneumoniae in vivo. Flow cytometric analysis of lung (Figure 4D) and BALF (Figure 4E) macrophages collected 12 hours after S. pneumoniae inoculation revealed that Ephx2–/– macrophages had reduced bacterial internalization compared with WT macrophages. We also observed the reduced internalization of S. pneumoniae by Ephx2–/– macrophages by immunofluorescence staining of lungs collected 12 hours after inoculation (Figure 4F). Ephx2 disruption similarly reduced phagocytosis by lung and BALF neutrophils in vivo (Supplemental Figure 5, A and B). In contrast, WT and Ephx2–/– macrophages displayed similar efferocytosis of dead lung cells, splenocytes, and thymocytes (Supplemental Figure 6). Thus, genetic disruption of Ephx2 impaired macrophage phagocytosis of S. pneumoniae, but not K. pneumonia or S. aureus. In contrast, Ephx2 disruption appeared to have little or no effect on macrophage efferocytosis of dead cells.
 Figure 4
Figure 4Defective phagocytosis of S. pneumoniae by Ephx2–/– macrophages in vitro. Phagocytosis of FITC-labeled K. pneumoniae (A), FITC-labeled S. pneumoniae (B), and Alexa Fluor 488–labeled S. aureus (C) by WT and Ephx2–/– peritoneal macrophages was determined by flow cytometry. Incubations were performed at a 10:1 ratio of bacteria/macrophages. n = 10 per group. *P < 0.05. Twelve hours after inoculation with 2 × 105 CFU/mouse S. pneumoniae, F4/80+Siglec F+CD11b+ macrophages were isolated from lungs (D) or BALF (E) of WT and Ephx2–/– mice. Phagocytosis of FITC-labeled S. pneumoniae was determined by flow cytometry. n = 5 per group. (F) Confocal images showing phagocytosis of S. pneumoniae by WT and Ephx2–/– macrophages (white arrows) 12 hours after inoculation. Sections were stained for S. pneumoniae (FITC), Ly6G (Violet 421), Siglec F (phycoerythrin), Ly6C (Violet 605), and DAPI and captured at ×100 magnification. Scale bars: 10 μm. Original magnification, ×40 (insets were digitally enlarged by 2.6-fold). *P < 0.05 by Student’s t test (A–C) or ordinary 2-way ANOVA, followed by Tukey’s post hoc multiple-comparison test (D and E).
Ephx2–/– macrophages have reduced cytokine and PRR expression after S. pneumoniae infection and following PGN stimulation. Macrophages are a major source of proinflammatory cytokines during bacterial infection. To determine whether sEH is involved in regulating macrophage cytokine production following infection, we measured cytokine mRNA levels in WT and Ephx2–/– peritoneal macrophages 4 hours after exposure to S. pneumoniae or PGN in vitro. We found that Ephx2–/– peritoneal macrophages had significantly lower Il1b, Il6, and Tnfa mRNA levels than did WT peritoneal macrophages after exposure to S. pneumoniae (Figure 5A). Both purified PGN and the synthetic TLR2 agonist Pam3CSK4 induced Il6 and Tnfa expression in peritoneal macrophages (Supplemental Figure 7A). Interestingly, PGN- and Pam3CSK4-induced cytokine expression was attenuated in Ephx2–/– peritoneal macrophages compared with WT peritoneal macrophages (Supplemental Figure 7, A and B).
 Figure 5
Figure 5Impaired proinflammatory cytokine responses to S. pneumoniae in Ephx2–/– macrophages in vitro. Peritoneal macrophages from WT and Ephx2–/– mice were stimulated with S. pneumoniae (1 × 106 CFU) for 4 hours. Macrophages were then assayed to measure the levels of Il1b, Il6, and Tnfa (A) and Pglyrp1, Tlr2, or Tlr4 (B) by real-time quantitative RT-PCR. Data represent the mean ± SEM. n = 6 per group. (C) Confocal microscopy of TLR2 in lung macrophages from WT and Ephx2–/– mice after S. pneumoniae infection in vivo. Scale bars: 10 μm. Original magnification, ×40 (insets were digitally enlarged by 2.6-fold). (D) Tnfa, Pglyrp1, and Tlr2 transcript levels, as measured by real-time quantitative RT-PCR, in WT macrophages treated or not with 10 μg/mL PGN in the presence or absence of 1 μM 11,12-EET, 1 μM 14,15-EET, or 10 μM 14,15-EEZE. n = 5 per group. *P < 0.05, by Student’s t test (A and B) or ordinary 1-way ANOVA, followed by Tukey’s post hoc multiple-comparison test (D).
PRRs play a key role in the early recognition of invading bacterial pathogens and in initiating the innate immune response. These receptors recognize Gram-positive and Gram-negative bacteria via conserved structures on the bacterial surface to activate the cell-signaling events that induce phagocytosis. Macrophages express TLR2 and PGLYRP1 receptors, which detect PGN on Gram-positive bacteria and activate phagocytosis (22, 42). To determine whether sEH regulates phagocytosis at the level of PRR expression, we measured mRNA levels of Pglyrp1, Tlr2, and Tlr4 in Ephx2–/– and WT peritoneal macrophages after exposure to S. pneumoniae or treatment with PGN. Ephx2–/– peritoneal macrophages had reduced expression of Tlr2 and Pglyrp1 compared with WT peritoneal macrophages after treatment with S. pneumoniae (Figure 5B) or PGN (Supplemental Figure 7C). In contrast, we observed that Tlr4 expression was not different between Ephx2–/– and WT peritoneal macrophages after S. pneumoniae (Figure 5B) or PGN treatment (Supplemental Figure 7B).
We also examined the expression of cytokines and PRRs in lung macrophages after treatment with PGN. The expression of Pglyrp1 was lower in Ephx2–/– lung macrophages compared with expression levels in WT lung macrophages after PGN stimulation (Supplemental Figure 8). In contrast, the expression of Pglyrp2, Pglyrp3, and Pglyrp4 was similar in PGN-stimulated WT and Ephx2–/– lung macrophages (Supplemental Figure 8). PGN-stimulated Ephx2–/– lung macrophages also had reduced proinflammatory cytokine and PRR expression compared with WT lung macrophages (Supplemental Figure 9). Confocal microscopy confirmed that, compared with WT lung macrophages, Ephx2–/– lung macrophages had reduced TLR2 expression in vivo following infection with S. pneumoniae (Figure 5C).
Ephx2 disruption probably regulates macrophage function via reduced epoxy fatty acid hydrolysis; therefore, we examined the effect of treatment with exogenous EETs or the putative EET receptor antagonist 14,15-EEZE on WT macrophage function. Importantly, both 11,12-EET and 14,15-EET inhibited Tnfa, Pglyrp1, and Tlr2 expression after PGN stimulation (Figure 5D). In contrast, treatment with 14,15-EEZE increased the expression of Tnfa, Pglyrp1, and Tlr2 after PGN stimulation (Figure 5D). Together, these observations suggest that sEH-mediated hydrolysis of epoxy fatty acids was required for maximal proinflammatory cytokine and PRR expression during S. pneumoniae infection or following PGN treatment.
Ephx2–/– macrophages exhibit reduced PGLYRP1 and TLR2 downstream signaling. The MAPKs, including ERK1/2, JNK1/2, and p38 MAPK, mediate many cellular functions including activation of various transcription factors such as NF-κB, production of pro- and antiinflammatory cytokines, and induction of phagocytosis in macrophages (43–45). To determine whether sEH regulates these downstream signaling events in macrophages, we compared phosphorylation of ERK, p38 MAPK, and IkBα (the endogenous repressor of NF-κB) in Ephx2–/– and WT peritoneal macrophages after treatment with PGN, S. pneumoniae, or LPS. Compared with WT macrophages, Ephx2–/– macrophages had significantly less phosphorylation of ERK, p38 MAPK, and IkBα after PGN and S. pneumoniae treatment (Figure 6, A–C). In contrast, we found no differences in ERK or p38 MAPK phosphorylation between Ephx2–/– and WT macrophages following LPS treatment (Figure 6D). The differences in response to PGN, but not LPS, were consistent with the observation that Ephx2–/– macrophages had impaired clearance of S. pneumoniae, but not K. pneumoniae.
 Figure 6
Figure 6Impaired signaling in Ephx2–/– macrophages after PGN treatment or infection with S. pneumoniae. (A and B) Immunoblot analysis of phosphorylated (p-) and total ERK, p38 MAPK, and IkBα in WT and Ephx2–/– macrophages stimulated with 10 μg/mL PGN for 0–60 minutes. (C) Immunoblot analysis of phosphorylated and total ERK in WT and Ephx2–/– macrophages infected with 1 × 106 CFU S. pneumoniae for 0–60 minutes. (D) Immunoblot analysis of phosphorylated and total ERK and p38 MAPK in WT and Ephx2–/– macrophages treated with 1 μg/mL LPS. (E) GTPase family proteins such as Cdc42 and Rac1 are inactive when bound to GDP and active when bound to GTP. Regulation of this molecular switch occurs through a GDP-GTP cycle that is controlled by the opposing activities of guanine nucleotide exchange factors (GEFs), which catalyze the exchange of GDP for GTP, and GTPase-activating proteins (GAPs), which increase the rate of GTP hydrolysis to GDP. GTPases interact with various effector proteins to influence their activity and/or localization, which ultimately affects macrophage phagocytosis. (F) GTP-Cdc42 and GTP-Rac1 levels were analyzed by immunoblotting in WT and Ephx2–/– macrophages stimulated with 10 μg/mL PGN for 0–60 minutes. For all immunoblots, data are representative of at least 3 independent experiments, and β-actin was used as a loading control.
The Rho family GTPases, including cell division control protein 42 homolog (Cdc42), Ras-related C3 botulinum toxin substrate 1 (Rac1), and RhoA, regulate cell shape, cell motility, and phagocytosis; in their GTP-bound active state, they interact with effectors that alter the actin cytoskeleton, contractility, and vesicle fusion (Figure 6E and ref. 46). Pathogen recognition receptor–mediated phagocytosis is a spatially and temporally regulated process that requires the functions of Rac1 and Cdc42 (47). To determine the role of Rac1 and Cdc42 in sEH-mediated macrophage phagocytosis of S. pneumoniae, we examined the activation of Rac1 and Cdc42 after PGN stimulation. Ephx2–/– macrophages had decreased active GTP-Rac1 and GTP-Cdc42 levels after PGN stimulation compared with WT macrophages (Figure 6F). Together, these data suggest that sEH regulated macrophage responses to PGN and S. pneumoniae through reduced phosphorylation of p38, ERK1/2, and IkBα, and impaired Rac1 and Cdc42 activation.
To determine whether the inability of Ephx2–/– macrophages to induce PGLYRP1 and TLR2 was directly related to their defects in downstream MAPK signaling and phagocytosis, we induced heterologous overexpression of PGLYRP1 and TLR2 (Supplemental Figure 10) and examined these outcomes in WT and Ephx2–/– macrophages. In WT macrophages, PGN induced the phosphorylation of ERK and p38 MAPK in a time-dependent manner with maximal effects at 10–30 minutes (Figure 7A). In contrast, Ephx2–/– macrophages were deficient in ERK and p38 MAPK activation. Importantly, heterologous overexpression of either PGLYRP1 or TLR2 in Ephx2–/– macrophages restored MAPK activation (Figure 7A) and increased the phagocytosis of FITC-labeled S. pneumoniae to WT levels (Figure 7B). Thus, PRR overexpression rescued the downstream MAPK signaling and phagocytosis phenotypes in Ephx2–/– macrophages.
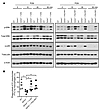 Figure 7
Figure 7TLR2 and PGLYRP1 overexpression rescue downstream signaling and phagocytosis defects in Ephx2–/– macrophages. (A) Immunoblot analysis of total and phosphorylated p38 MAPK and ERK in PGN-stimulated WT macrophages, Ephx2–/– macrophages, and Ephx2–/– macrophages overexpressing either TLR2 or PGLYRP1. n = 5 per group. β-Actin was used as a loading control. (B) Phagocytosis of FITC-labeled S. pneumoniae by WT macrophages, Ephx2–/– macrophages and Ephx2–/– macrophages overexpressing either TLR2 or PGLYRP1 as measured by flow cytometry. Each square represents an individual mouse: white squares, WT; black squares, Ephx2–/–). n = 5 per group. *P < 0.05, by ordinary 2-way ANOVA, followed by Tukey’s post hoc multiple-comparison test (B).
TLR2–/– and PGLYRP1–/– macrophages have reduced S. pneumoniae phagocytosis and PGN-stimulated cytokine induction. PGLYRP1 is a known receptor for S. pneumoniae; however, it is unknown whether PGLYRP1 also regulates macrophage phagocytosis and/or proinflammatory cytokine production. To address this question, we obtained peritoneal macrophages from Tlr2–/–, Pglyrp1–/–, and WT littermate control mice and examined macrophage phagocytosis and proinflammatory cytokine production after PGN stimulation, with or without EET treatment. We found that both Tlr2 and Pglyrp1 disruption significantly impaired macrophage phagocytosis of S. pneumoniae and attenuated PGN-stimulated Tnfa, Il1b, and Il6 induction (Figure 8, A and B, and Supplemental Figure 11). Compared with the vehicle-treated controls, EET treatment significantly impaired phagocytosis by WT macrophages; however, EETs did not further suppress phagocytosis by either Tlr2–/– or Pglyrp1–/– macrophages (Figure 8A). Similarly, EET treatment significantly suppressed the induction of Tnfa, Il1b, and Il6 in PGN-stimulated WT macrophages; however, EETs did not further attenuate cytokine induction beyond the suppression observed in the vehicle-treated Tlr2–/– or Pglyrp1–/– macrophages (Figure 8B and Supplemental Figure 11).
 Figure 8
Figure 8Tlr2–/– and Pglyrp1–/– macrophages have reduced S. pneumoniae phagocytosis and PGN-stimulated cytokine induction. (A) Peritoneal macrophages were isolated from Tlr2–/– or Plglyrp1–/– mice and treated for 4 hours with vehicle, 1 μM 11,12-EET, or 1 μM 14,15-EET. Phagocytosis of FITC-labeled S. pneumoniae was determined by flow cytometry. (B) Peritoneal macrophages were isolated from Tlr2–/– or Plglyrp1–/– mice and treated for 4 hours with PBS or 10 μg/mL PGN in the presence of vehicle, 1 μM 11,12-EET, or 1 μM 14,15-EET. Expression of Tnfa was determined by real-time quantitative RT-PCR. n = 9 per group. (C) Peritoneal macrophages were isolated from Tlr2–/– or Plglyrp1–/– mice and treated for 4 hours with 10 μg/mL PGN (Control) or conditioned medium from WT PGN-treated macrophages (WT Cond Media), and phagocytosis of FITC-labeled S. pneumoniae was determined by flow cytometry. n = 5 per group. *P < 0.05, by ordinary 2-way ANOVA, followed by Tukey’s post hoc multiple-comparison test (A–C).
These data beg the question as to whether reduced phagocytosis in PRR-deficient mice is due to a lack of PGLYRP1 or TLR2 expression or to diminished cytokine induction by Tlr2–/– or Pglyrp1–/– macrophages. To distinguish between these possibilities, we attempted to rescue S. pneumoniae phagocytosis in Tlr2–/– and Pglyrp1–/– macrophages by priming them with conditioned media from PGN-treated WT macrophages. Importantly, we found that the conditioned medium failed to restore S. pneumoniae phagocytosis by Tlr2–/– or Pglyrp1–/– macrophages (Figure 8C). Together, these data suggest that sEH deficiency or EET treatment regulates macrophage phagocytic and inflammatory responses to PGN and S. pneumoniae mainly through suppression of TLR2 and PGLYRP1 expression.
EETs inhibit proinflammatory cytokine production, PRR expression, and phagocytosis of S. pneumoniae in human macrophages. To determine whether EETs can also regulate human proinflammatory cytokine production, PRR expression, and macrophage phagocytosis, we examined the effects of exogenous EETs on these endpoints in peripheral blood monocyte–derived macrophages and isolated alveolar macrophages from healthy volunteers. Both 11,12-EET and 14,15-EET significantly reduced phagocytosis of FITC-labeled S. pneumoniae by PGN-treated human monocyte–derived macrophages (Figure 9A). Treatment with EETs also attenuated the induction of IL1B, IL6, TNFA, and TLR2 mRNAs in PGN-stimulated monocyte-derived macrophages (Figure 9A). In addition, both 11,12-EET and 14,15-EET reduced human alveolar macrophage phagocytosis of S. pneumonia (Figure 9B), but not K. pneumoniae (Supplemental Figure 12A). EETs also attenuated the induction of IL6, TNFA, TLR2, and PGLYRP1 in human alveolar macrophages stimulated with PGN (Figure 9B), but not with LPS (Supplemental Figure 12, B and C). Together, these results suggest that EETs have inhibitory effects on phagocytosis, proinflammatory cytokine production, and PRR expression in human macrophages, like in mouse macrophages.
 Figure 9
Figure 9EETs inhibit innate immune responses of human macrophages. (A) Phagocytosis of FITC-labeled S. pneumoniae (measured by flow cytometry) and expression of IL1B, IL6, TNFA, and TLR2 transcripts (measured by real-time quantitative RT-PCR) in human monocyte–derived macrophages treated or not with 10 μg/mL PGN in the presence or absence of 1 μM 11,12-EET or 14,15-EET. (B) Phagocytosis of FITC-labeled S. pneumoniae (measured by flow cytometry) and expression of IL1B, IL6, TNFA, TLR2, and PGLYRP1 transcripts (measured by real-time quantitative RT-PCR) in human AMs treated or not with 10 μg/mL PGN in the presence or absence of 1 μM 11,12-EET or 1 μM 14,15-EET. n = 5–15 per group. *P < 0.05, by repeated-measures ANOVA (phagocytosis panels in A and B) or ordinary 1-way ANOVA, followed by Tukey’s post hoc multiple-comparison test (remaining panels in A and B).
-
Discussion
The effect of sEH and EETs on innate immune responses of macrophages to bacterial pathogens had not to our knowledge been previously investigated. This study demonstrates that sEH regulation of EET levels in macrophages was critical for induction of the proinflammatory responses that were required for optimal phagocytosis of S. pneumoniae, but not S. aureus or K. pneumoniae. Specifically, our major findings are that (a) genetic disruption of Ephx2 or sEH inhibition in mice diminished lung clearance of S. pneumoniae in vivo; (b) Ephx2–/– macrophages exhibited diminished phagocytosis of S. pneumoniae in vitro; (c) exposure to the S. pneumoniae bacterial coat component PGN induced sEH expression and increased EET levels; (d) Ephx2 disruption suppressed the activation of MAPKs, NF-κB, and Rho family GTPases; (e) Ephx2 disruption or EET treatment attenuated the induction of proinflammatory cytokines and the PGN receptors TLR2 and PGLYRP1; (f) a putative EET receptor antagonist increased the expression of proinflammatory cytokines and PGN receptors and improved bacterial clearance in vivo; and (g) the role of EETs in regulating macrophage phagocytosis was well conserved between mice and humans. Together, these findings suggest that induction of sEH expression by PGN plays a critical role in phagocytosis and lung clearance of S. pneumoniae during acute infection and provide a host-centered approach to the treatment of S. pneumoniae bacterial infections in humans.
Ephx2–/– lungs had impaired clearance of the Gram-positive bacteria S. pneumoniae, whereas clearance of another Gram-positive bacteria, S. aureus, and clearance of the Gram-negative bacteria K. pneumoniae were unaffected. A previous study showed that knockout of sEH results in decreased neutrophil infiltration into the lungs in response to bacterial LPS (33). In contrast, our study showed that impaired clearance of S. pneumoniae in Ephx2–/– lungs was associated with increased numbers of neutrophils in the BALF, without changes in macrophage numbers. However, our studies revealed that Ephx2–/– macrophages produced more EETs and had reduced phagocytic ability. Similarly, treatment with exogenous EETs decreased macrophage phagocytosis of S. pneumoniae, and treatment of mice with a putative EET receptor antagonist increased the clearance of S. pneumoniae in vivo. Interestingly, EETs did not indiscriminately inhibit bacterial phagocytosis. Indeed, Ephx2 disruption did not alter K. pneumoniae or S. aureus phagocytosis or clearance, and others have shown that treatment of THP-1 monocytes with 11,12-EET increases phagocytosis of L. monocytogenes (48). It is unclear why Ephx2 disruption did not alter S. aureus phagocytosis, although S. aureus is rapidly cleared by mice and may be a less suitable model for bacterial pneumonia in humans (49). The varied responses may be related to differential PAMP expression or accessibility on the surfaces of different pathogens that activate specific cell-surface receptors and induce unique downstream signaling cascades that impact phagocytosis differently.
In this study, the S. pneumonia PAMP PGN induced sEH expression in WT mouse macrophages. This increase in sEH expression would be expected to reduce EET levels and attenuate their antiinflammatory effects during the acute phase of inflammation. Previous studies have shown that vascular sEH expression is regulated by both endogenous and exogenous factors during inflammation. Both homocysteine (50) and angiotensin II (51) are thought to enhance endothelial inflammation by increasing sEH expression and lowering EET levels. This inflammation appears to play a key role in atherosclerosis, as genetic disruption or pharmacological inhibition of sEH reduces neointimal plaque formation (52). EETs also inhibit VCAM-1 expression in response to TNF-α, IL-1β, or LPS in vitro (27). Conversely, lungs from LPS-treated Ephx2–/– mice show attenuated induction of cell adhesion molecule expression, reduced levels of chemokines, and decreased neutrophil infiltration in vivo (33). In most of these studies, inhibition of sEH resulted in a reduced inflammatory response that was thought to be beneficial. Indeed, sEH inhibitors have been developed and are being tested as novel treatments for a myriad of inflammatory diseases (53). Our study suggests that one potential pitfall of sEH inhibition is reduced bacterial phagocytosis by macrophages, which can lead to impaired bacterial clearance and result in uncontrolled infection and increased inflammation.
While sEH is often increased during inflammation, several studies suggest that CYP epoxygenase expression is decreased in monocytic cells during acute inflammation. CYP epoxygenase expression is downregulated early during acute inflammation (54, 55), which would reduce antiinflammatory EET production. EETs inhibit TNF-α secretion from THP-1 cells (56). Similarly, 11,12-EET inhibits LPS-mediated induction of COX-2 expression and PGE2 formation (57). Thus, monocytes appear to be programmed to decrease CYP epoxygenase expression and increase sEH expression during acute inflammation. Together, these changes result in reduced EET levels, attenuation of their antiinflammatory effects, and potentiation of the inflammatory response.
Binding of PGN to TLR2 in macrophages induces recruitment and activation of PI3K at the plasma membrane (58). PI3K induces subsequent activation of Rac1 and MAPKs such as ERK (58, 59). We found that Ephx2–/– or EET-treated macrophages had attenuated activation of ERK, p38 MAPK, and Rho family proteins by S. pneumoniae or PGN. The precise molecular mechanisms by which EETs suppress macrophage activation remain unclear. EETs are believed to signal through a high-affinity cell-surface GPCR and are known PPAR agonists (37). Although the receptor that mediates EET actions in vivo has not been identified, EETs are known to activate signaling of both Gαs and Gα12/13 (37, 60). Induction of Gαs/cAMP/protein kinase A signaling can impair both ERK and Rho family activation (61, 62). EETs may also have a direct effect on inflammatory receptor signaling, as agonists that induce Gαs/cAMP can inhibit PGN/TLR2 signaling (63). Alternatively, reduced ERK, p38 MAPK, and Rho family activation may be secondary to the lack of TLR2 and PGLYRP1 induction. Attenuation of ERK, p38 MAPK, and Rac1 activation by EETs critically regulates macrophage phagocytosis in 2 ways: (a) suppression of ERK, p38 MAPK, and Rac1 signaling reduces NF-κB activation and proinflammatory cytokine and receptor expression (58, 59); and (b) inhibition of Rac1 and Cdc42 activation blocks the cytoskeletal rearrangements and membrane remodeling necessary for phagocytosis (64). Thus, attenuation of PGN-induced activation of ERK, P38 MAPK, Rac1, and Cdc42 by EETs potently limited macrophage phagocytosis and clearance of S. pneumoniae.
Pglyrp1 and Tlr2 induction was selectively attenuated in Ephx2–/– macrophages after PGN and S. pneumoniae stimulation. Isolated Pglyrp1- and Tlr2-deficient macrophages were defective in macrophage activation, proinflammatory cytokine production, and effective phagocytosis of S. pneumoniae. Importantly, treatment of Pglyrp1- and Tlr2-deficient macrophages with exogenous EETs did not further reduce cytokine expression or S. pneumoniae phagocytosis. Conversely, heterologous overexpression of either PGLYRP1 or TLR2 in Ephx2–/– macrophages fully restored activation, inflammatory signaling, and phagocytosis. Therefore, a critical step in sEH regulation of macrophage phagocytosis of S. pneumoniae was at the level of PGN receptor expression. Our findings are consistent with the recent report that TLR2-deficient macrophages exhibit impaired phagocytosis and bacterial killing (65). The effect of EET treatment or Ephx2 disruption on PGN-induced receptor expression and inflammatory signaling appeared selective; Ephx2–/– macrophages did not have altered TLR4 expression after LPS treatment or altered phagocytosis of K. pneumoniae. Consistent with this finding, both Ephx2–/– and WT macrophages were activated and produced proinflammatory cytokines in response to LPS; however, only Ephx2–/– macrophages had impaired proinflammatory cytokine generation after PGN or S. pneumoniae treatment. While EETs are broadly characterized as antiinflammatory (66), these effects are indeed context dependent. In this study, the reduced inflammatory response to S. pneumonia PAMPs impaired the clearance of S. pneumoniae from Ephx2–/– lungs. Ultimately, Ephx2–/– mice experienced an increased bacterial load, prolonged infection, and increased inflammation.
A schematic representation of the effects of sEH and EETs on inflammatory signaling and phagocytosis of S. pneumoniae by macrophages is shown in Figure 10. In WT macrophages, after encountering PGN on S. pneumoniae, TLR2 and PGLYRP1 initiate the phosphorylation of ERK, p38 MAPK, and IκBα. This phosphorylation leads to activation of Rho family GTPases and the upregulation of proinflammatory cytokines and PRRs. PGN induces sEH expression and increased sEH-mediated hydrolysis of EETs, which potentiates the acute inflammatory response and induces bacterial phagocytosis. In the absence of sEH, increased EETs blunt the signaling cascades required for macrophage activation, dramatically reducing phagocytosis and bacterial clearance. Precisely how EETs inhibit TLR2 and PGLYRP1 signaling is not known. As with MAPK signaling, the identity of the receptor responsible for nuclear signaling events downstream of EETs remains enigmatic. EETs have been shown to regulate ATF6 and SP1 transcription factors, which are critical for TLR2 expression in monocytes and macrophages (67, 68). Identification of the putative EET receptor and delineation of EET-mediated signaling events that regulate TLR2 and PGLYRP1 expression will undoubtedly shed light on the role of EET and sEH in regulating the innate immune response to bacterial pathogens.
 Figure 10
Figure 10Proposed model for the regulation of bacterial clearance by EETs. In WT mice, PAMPs from S. pneumoniae (PGN) are recognized by PRRs (TLR2 and PGLYRP1), and this leads to upregulation of sEH, resulting in reduced EET levels. Low EETs permit enhanced signaling through ERK, p38 MAPK, and NF-κB and lead to the production of proinflammatory cytokines (IL-1, IL-6, TNF-α) and increased expression of PRRs. A positive feedback loop further enhances downstream signaling and ultimately leads to efficient activation of Rho family GTPases (Rac1 and Cdc42), which are responsible for enhanced bacterial phagocytosis and clearance. In Ephx2–/– mice, PAMPs cannot upregulate sEH, and this results in reduced EET hydrolysis and increased EET levels. Increased EETs attenuate downstream signaling through ERK, p38 MAPK, and NF-κB, reduce proinflammatory cytokine production, reduce the expression of PRRs, and diminish activation of the Rho family GTPases, resulting in impaired bacterial phagocytosis and clearance.
Since EETs regulate bacterial phagocytosis, genetic polymorphisms that result in altered EET formation and/or hydrolysis may predispose some patients to more severe or chronic infections. CYP2C8, CYP2C9, and CYP2J2 are responsible for the majority of EET formation in humans (69). Interestingly, nearly all known polymorphisms in these 3 genes lower the expression or activity of the respective P450 epoxygenases and increase cardiovascular disease risk (69–71). In contrast, our data suggest that these P450 polymorphisms may be protective in the setting of bacterial infection, as reduced EET production might result in more efficient bacterial clearance. There are 2 common polymorphisms in the EPHX2 gene that encode sEH proteins with altered activity: the K55R polymorphism results in increased EET hydrolysis activity, whereas the R287Q polymorphism results in decreased sEH dimerization and activity (72, 73). These sEH polymorphisms may be protective or detrimental to bacterial clearance in humans. Microsomal epoxide hydrolase/epoxide hydrolase 1 (mEH/EPHX1) also regulates EET levels in vivo (74). Thus, EPHX1 polymorphisms that increase or decrease mEH activity (75, 76) may also play a role in bacterial clearance. The genetics that underlie EET metabolism may have a significant, but underappreciated, role in the enormous public health burden of S. pneumoniae infections. In addition, the use of EEZE or other EET receptor antagonists could offer clinical benefit for patients with severe or refractory pneumonia.
In summary, we found that phagocytosis of S. pneumoniae was severely impaired in Ephx2–/– mice in vivo. The induction of sEH by PGN and subsequent reduction of EETs were critical to the regulation of the innate immune response and optimal clearance of S. pneumoniae from the lungs. Mechanistically, these effects were primarily due to EET-mediated downregulation of TLR2 and PGLYRP1 expression, diminished production of proinflammatory cytokines, and reduced activation of p38 MAPK, ERK1/2, IkBα, Rac1/2, and Cdc42. The role of sEH in bacterial clearance may become particularly important should sEH inhibitors be approved for the treatment for cardiovascular disease in humans. Conversely, the improvement in bacterial clearance we observed after treatment with EET antagonists such as EEZE highlights a potential new approach for the treatment of bacterial pneumonia that targets the host rather than the pathogen.
-
Methods
Mice. Male WT C57BL/6J and Tlr2–/– mice (B6.129-Tlr2tm1Kir/J; stock no. 004650) were obtained from The Jackson Laboratory. Ephx2–/– mice and their WT littermate controls were generated by breeding Ephx2+/– Ephx2+/– mice on a pure C57BL/6 genetic background at the NIEHS. Pglyrp1–/– mice were a gift from Roman Dziarski (Indiana University School of Medicine, Indianapolis, Indiana, USA). Mice were genotyped using previously published PCR-based methods (32) and used at 8–12 weeks of age.
Reagents and bacterial strains. The following reagents were purchased from MilliporeSigma: FITC, LPS from Salmonella typhosa or E. coli, LTA from S. aureus, Zym from Saccharomyces cerevisiae, and Man from S. cerevisiae. PGN from S. aureus was purchased from Fluka Chemical. The synthetic lipoprotein Pam3CSK4 (TLR2 agonist) was purchased from InvivoGen. Antibodies against ERK (catalog 9101), p-ERK (catalog 4695), p38 MAPK (catalog 9212), pp38 MAPK (catalog 9211), IkBα (catalog 4812), and pIkBα (catalog 2859) were purchased from Cell Signaling Technology. Antibodies against sEH (sc-22344) were purchased from Santa Cruz Biotechnology. Antibodies against β-actin (A2228) were purchased from MilliporeSigma. The Rac1-, Cdc42-pulldown kit (17-10394) was purchased from MilliporeSigma. K. pneumoniae 43816 (serotype 2), S. pneumoniae 6303 (serotype 3), and S. aureus BAA-1717 (USA 300-Hou-MR) were purchased from the American Type Culture Collection (ATCC). DMEM was purchased from Life Technologies (Thermo Fisher Scientific), and FBS was purchased from Gemini. Bacterial cultural plates containing glucose mineral salt and trypticase soy agar (with 5% sheep blood) were made at the NIEHS. Synthetic EETs and 14,15-EEZE were purchased from Cayman Chemical Company. TPPU [1-trifluoromethyloxyphenyl-3-(1-propionylpiperidin-4-yl) urea] was provided by Bruce Hammock (UC Davis, Davis, California, USA).
In vivo bacterial infection. Male Ephx2–/– mice and age-matched WT controls were infected with either 1 × 103 or 2 × 103 CFU/50 μL K. pneumoniae, 2 × 105 CFU/50 μL S. pneumoniae, or 3.3 × 107 or 6.6 × 107 CFU/50 μL S. aureus as indicated. Bacteria were administered to the lungs by intranasal instillation under isoflurane anesthesia. In some experiments, the sEH inhibitor TPPU was solubilized in PEG400 and added to the drinking water for a final concentration of 10 mg/L TPPU and 1% PEG400. Mice were given a TPPU dose of approximately 1.7 mg/kg/day and 1% PEG400 in their drinking water for 6 days prior to infection with S. pneumoniae. In other studies, mice were given 15 μg/kg/day 14,15-EE-5(Z)-E administered via subcutaneously implanted osmotic minipumps (1007D; Alzet) 4 days prior to infection with S. pneumoniae. Twelve, 24, or 48 hours after instillation, the lungs were removed and homogenized in 1 mL sterile PBS. The homogenized lung samples were serially diluted and plated on the appropriate agar plates to count CFU to determine bacterial clearance from the lungs. The bacterial doses used for in vivo infection were determined by plating a fraction of the inoculum prior to infection. For some experiments, lung tissues were fixed in formalin, sectioned (5 μm), and stained with H&E. A pathologist who was blinded to treatment group assignment and genotype scored the percentage of the lung showing inflammation on a scale of 0 to 4 (0 = 0%; 1 = 1%–10%; 2 = 11%–30%; 3 = 31%–50%; 4 = >50%).
BALF collection and analysis. BALF was collected immediately after euthanasia, and cell counts were performed as previously described (77). Differential analysis of BALF cells was performed according to standard procedures by counting at least 100 cells.
RNA isolation and real-time quantitative reverse transcriptase PCR. RNA was isolated using the RNeasy kit (QIAGEN). cDNA was generated from 1.5 μg purified RNA using reverse transcription reagents from Applied Biosystems. Real-time quantitative reverse transcriptase PCR (RT-PCR) was performed in triplicate using Maxima qPCR Master Mix (Thermo Fisher Scientific) in the HT7900 ABI Sequence Detection System (Applied Biosystems). TaqMan-based primer/probe assays for IL1B, IL6, TNFA, TLR2, TLR4, Pglyrp1, Pglyrp2, Pglyrp3, Pglyrp4, and Ephx2 were purchased from Applied Biosystems. Gene expression was normalized to Gapdh, and expression levels in untreated (control) samples were set at 1.
Analysis of macrophage function. Peritoneal macrophages were collected from groups of 3–5 WT or Ephx2–/– mice 45 days after injection of 1 mL 3% Brewer’s thioglycolate broth. Cells were incubated at 37°C for 30 minutes, and the adherent cells were collected and determined to be 85%–90% Mac-1+ by flow cytometric analysis. Macrophages were stimulated (1 × 106 cells/mL; 0.5 mL/well) in 24-well plates for 4 hours with PAMPs (10 μg/mL unless otherwise noted). For in vitro bacterial stimulation, macrophages were seeded at a density of 2 × 105 cells/well on 24-well plates in antibiotic-free medium. Four hours later, heat-killed (control) or live S. pneumoniae (1 × 106 CFU) were added to the plates. The cells were collected for preparation of RNA and real-time quantitative RT-PCR analysis. For analysis of cell signaling, whole-cell lysates from peritoneal macrophages stimulated with PGN (10 μg/mL) or 1 × 106 CFU S. pneumoniae were immunoblotted with antibodies against phosphorylated and total p38 MAPK, IκBα, ERK1/2, and β-actin. Macrophage phagocytosis assays were performed according to previously published protocols (48). Briefly, S. pneumoniae and K. pneumoniae were heat killed by boiling for 10 minutes and conjugated with FITC using a microlabeling kit (Thermo Fisher Scientific). S. aureus were heat killed by boiling for 10 minutes and conjugated with Alexa Fluor 488 using a microlabeling kit (Thermo Fisher Scientific). Isolated macrophages were activated with 1 μg/mL PGN for 48 hours and then mixed with bacteria (FITC–S. pneumoniae, FITC–K. pneumoniae, and Alexa Fluor 488–S. aureus) in a suspension at a ratio of 1:10 (cell/bacteria), rotated at 37°C for 20 minutes, washed 3 times with PBS, and analyzed by flow cytometry on an LSR II (BD). At least 200 macrophages per sample were examined using an LSM 710 confocal microscope (Zeiss). The phagocytic index was calculated as the percentage of macrophages containing at least 1 bacterium times the mean number of bacteria per positive cell. For some experiments, lung macrophages from WT and Ephx2–/– mice were sorted by flow cytometry using CD11b+ and F4/80+ markers and used in place of peritoneal macrophages. For other experiments, macrophages were incubated with synthetic EETs (1 μM each) or the selective sEH inhibitor t-AUCB (1 μM).
Confocal microscopy. Frozen lung sections were fixed in methanol with 0.3% H2O2 at 4°C, permeabilized with Triton X-100 (0.8%), and stained with the fluorescent antibodies: FITC–rabbit anti–S. pneumoniae (MyBioSource.com, MBS324034); PE–anti–mouse Siglec F (BioLegend, catalog 155506); Brilliant Violet 421–anti–mouse Ly-6G (BioLegend, catalog 127628); and Brilliant Violet 605–anti–mouse Ly-6C (BioLegend, catalog 128038). Separate slides were stained with FITC–anti–mouse/human CD282 (TLR2) (BioLegend, catalog 121085); Alexa Fluor 594 anti–mouse/human CD11b antibody (BioLegend, catalog 101254); and DAPI.
Protein immunoblotting. Macrophages were lysed in 1× Laemmli with 20 mM DTT. Proteins were resolved on a 10% SDS-PAGE gel, transferred onto nitrocellulose membranes (Thermo Fisher Scientific), and probed with primary antibodies (all used at 1:1000). Membranes were then washed in 0.1% Tween PBS (TPBS) and exposed for 60 minutes to 1:5000 species-specific HRP-conjugated secondary antibody (Calbiochem) in 1% BSA with TPBS. Signal was detected with ECL Western Blot Detection Reagents (GE Healthcare) followed by film exposure (GE Healthcare).
Knockdown of the Pglyrp1 receptor using siRNA. Freshly prepared peritoneal macrophages were transfected with 20 nmol siRNA targeted to the Pglyrp1 receptor, negative control siRNA, or GFP control vector using Lipofectmin 2000 (Amaxa). Transfected cells were then stimulated with 10 μg/mL PGN as described above and analyzed by real-time quantitative RT-PCR after 4 hours. The following siRNAs were purchased from MilliporeSigma: (siRNA-1) SASI_Mm01_00062898 and SASI_Mm01_00062898_AS; (siRNA-2) SASI_Mm01_00062897 and SASI_Mm01_00062897_AS; and (siRNA-3) SASI_Mm01_00062895 and SASI_Mm01_00062895 _AS.
Analysis of Pglyrp1–/– and Tlr2–/– macrophage function after priming with WT conditioned medium. Peritoneal macrophages were collected from groups of 3–5 Pglyrp1+/+, Tlr2+/+, Pglyrp1–/–, and Tlr2–/– mice 5 days after injection of 1 mL 3% thioglycolate broth. Cells were incubated at 37°C for 30 minutes, and the adherent cells were collected and determined to be 85%–90% Mac-1+ by flow cytometric analysis. Macrophages were stimulated (1 × 106 cells/mL; 0.5 mL/well) in 24-well plates for 4 hours with PBS, 10 μg/mL PGN, vehicle (ethanol), and 1 μM 11,12-EET or 14,15-EET before collection for flow cytometric determination of FITC-labeled S. pneumoniae phagocytosis or mRNA expression analyses. In some experiments, macrophages were primed for 4 hours using conditioned medium from WT macrophages that had been incubated with 10 μg/mL PGN for 4 hours. Primed macrophages were assessed for phagocytosis by incubation with 1 × 106 FITC S. pneumoniae in suspension at a ratio of 1:10 (cell/bacteria), rotated at 37°C for 20 minutes, washed 3 times with PBS, and analyzed by flow cytometry on an LSR II (BD).
Measurement of endogenous EETs. EETs in media were analyzed by LC-MS/MS. Media were acidified to 0.05% acetic acid in 2.5% methanol (final volumes), spiked with the internal standard (3 ng 11,12-EET-d11) (Cayman Chemical), and passed through HyperSep Retain SPE columns (Thermo Fisher Scientific). The columns were washed with 0.05% acetic acid in 2.5% methanol and eluted with 0.5 mL methanol and 1 mL ethyl acetate. Samples were dried in a vacuum centrifuge, resuspended in 50 μL of 30% ethanol, and injected into the Agilent 1200 Series capillary HPLC (Agilent Technologies) coupled to an API 3000 triple-quadrupole mass spectrometer (PE SCIEX) with negative mode electrospray ionization and multiple reaction monitoring as previously described (78).
Vectors, virus production, and Tlr2 and Pglyrp1 expression. Tlr2 and Pglyrp2 were overexpressed in macrophages using the doxycycline-inducible p-INDUCER 20 lentiviral vector system (79). The pINDUCER20 lentiviral vector was a gift from Guang Hu (NIEHS, NIH). Coding sequences for mouse Tlr2 and Pglyrp1 were PCR amplified from cDNA isolated from macrophages treated with PGN using the following specific primers for Tlr2 and Pglyrp1: Tlr2 forward, 5′-CACCATGCTACGAGCTCTTTGGCTCTT-3′, Tlr2 reverse, 5′-CACCCTAGGACTTTATTGCAGTTCTCAGATTT-3′; Pglyrp1 forward, 5′-CACCATGTTGTTTGCCTGTGCTCT-3′, Pglyrp1 reverse, 5′-CACCTCACTCTCGGTAGTGTTCCCA-3′. pINDUCER20 was digested with Xba I and BamH I followed by insertion of Tlr2 and Pglyrp1 to construct the mouse Tlr2-pINDUCER20 and Pglyrp1-pINDUCER20 plasmids, respectively. Mouse Tlr2-pINDUCER20 and Pglyrp1-pINDUCER20 plasmids were packaged in lentivirus and concentrated by the NIEHS Virus Vector Core.
Human macrophage experiments. Human peripheral blood samples and human BALF samples were collected at the NIEHS Clinical Research Unit. Patients were excluded if they had received cytotoxic drugs in the previous 3 months or had taken steroid or nonsteroidal antiinflammatory medications within 72 hours of sample collection. Human peripheral blood monocytes were differentiated into macrophages by treatment with GM-CSF for 5–7 days (80). Human AMs were collected by bronchoscopy with bronchoalveolar lavage and used directly to examine their innate immune responses.
Statistics. All data are presented as the mean ± SEM. Statistical comparisons between treatment groups were performed using an ordinary 2-way ANOVA followed by post hoc Tukey’s multiple-comparison test or by an unpaired Student’s t test for 2 groups using GraphPad Prism (GraphPad Software). Statistical significance was defined as a P value of less than 0.05.
Study approval. The handling and care of the animals as well as all animal procedures were conducted in conformity with the NIH’s Guide for the Care and Use of Laboratory Animals (National Academies Press, 1996). All experiments were approved by the NIEHS Animal Care and Use Committee. Human peripheral blood collection was done under protocol 10-E-0063, which was approved by the NIEHS IRB. Informed consent was obtained from each healthy volunteer.
-
Author contributions
HL, MLE, and DCZ conceptualized the project and wrote the manuscript. HL, JAB, MLE, JC, SLH, and LMG performed research and analyzed data. AG and JPG designed and generated expression constructs. MBF, SG, and SHS obtained human macrophages for analysis. The order of the co–first authors was determined on the basis of their contributions to this project. HL conceptualized the project, provided critical immunological guidance, jointly performed most of the experiments, and wrote the initial manuscript. JAB introduced and optimized novel models and methods for the project and co-performed nearly all of the experiments. MLE conceptualized the project, performed multiple analyses, supervised the research, and wrote and edited the manuscript. All authors edited the manuscript and approved the final version.
-
Acknowledgments
The authors would like to thank Roman Dziarski (Indiana University School of Medicine–Northwest) for providing Pglyrp1–/– mice and Bruce Hammock (UC Davis) for providing TPPU. We also thank Jeff Tucker of the NIEHS Microscopy Core; Carl Bortner and Maria Sifre of the NIEHS Flow Cytometry Core; Negin Martin and David Chen of the NIEHS Viral Vector Core; and Debra King of the NIEHS Clinical Pathology Group for their support of this study. This work was supported by the Intramural Research Program of the NIH, NIEHS (Z01 ES025034, to DCZ).
Address correspondence to: Darryl C. Zeldin, Division of Intramural Research, NIH/NIEHS, 111 T.W. Alexander Drive, Building 101, Room A214, Research Triangle Park, North Carolina 27709, USA. Phone: 984.287.3641; Email: zeldin@niehs.nih.gov.
-
Footnotes
Conflict of interest: DCZ is a co-inventor on several patents (US patents 6,531,506: “Inhibitors of Epoxide Hydrolases for the Treatment of Hypertension;” 6,693,130: “Inhibitors of Epoxide Hydrolases for the Treatment of Hypertension;” and 6,916,843: “Anti-inflammatory Actions of Cytochrome P450 Epoxygenase-Derived Eicosanoids”) for the use of epoxide hydrolase inhibitors for the treatment of inflammation and cardiovascular diseases.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(22):e129679.https://doi.org/10.1172/JCI129679.
-
References
- Mandell LA, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44(Suppl 2):S27–S72.View this article via: PubMed Google Scholar
- Levine OS, et al. Pneumococcal vaccination in developing countries. Lancet. 2006;367(9526):1880–1882.
- Kadioglu A, Andrew PW. The innate immune response to pneumococcal lung infection: the untold story. Trends Immunol. 2004;25(3):143–149.
- Bosso JA, Drew RH. Application of antimicrobial stewardship to optimise management of community acquired pneumonia. Int J Clin Pract. 2011;65(7):775–783.
- Smith AM, et al. Mathematical model of a three-stage innate immune response to a pneumococcal lung infection. J Theor Biol. 2011;276(1):106–116.
- Marriott HM, Dockrell DH. The role of the macrophage in lung disease mediated by bacteria. Exp Lung Res. 2007;33(10):493–505.
- Pittet LA, et al. Earliest innate immune responses require macrophage RelA during pneumococcal pneumonia. Am J Respir Cell Mol Biol. 2011;45(3):573–581.
- Serhan CN, et al. Resolution of inflammation: state of the art, definitions and terms. FASEB J. 2007;21(2):325–332.
- Mancuso P, et al. Intrapulmonary administration of leukotriene B4 enhances pulmonary host defense against pneumococcal pneumonia. Infect Immun. 2010;78(5):2264–2271.
- Aronoff DM, et al. Prostaglandin E2 inhibits alveolar macrophage phagocytosis through an E-prostanoid 2 receptor-mediated increase in intracellular cyclic AMP. J Immunol. 2004;173(1):559–565.
- Aronoff DM, et al. Synthetic prostacyclin analogs differentially regulate macrophage function via distinct analog-receptor binding specificities. J Immunol. 2007;178(3):1628–1634.
- Hoffmann JA, et al. Phylogenetic perspectives in innate immunity. Science. 1999;284(5418):1313–1318.
- Crouch E, Wright JR. Surfactant proteins a and d and pulmonary host defense. Annu Rev Physiol. 2001;63:521–554.
- Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1(2):135–145.
- Takeuchi O, et al. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11(4):443–451.
- Alexopoulou L, et al. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413(6857):732–738.
- Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282(5396):2085–2088.
- Hayashi F, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410(6832):1099–1103.
- Takeuchi O, et al. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int Immunol. 2001;13(7):933–940.
- Heil F, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303(5663):1526–1529.
- Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408(6813):740–745.
- Dziarski R. Peptidoglycan recognition proteins (PGRPs). Mol Immunol. 2004;40(12):877–886.
- Lee MH, et al. Peptidoglycan recognition proteins involved in 1,3-beta-D-glucan-dependent prophenoloxidase activation system of insect. J Biol Chem. 2004;279(5):3218–3227.
- Liu C, et al. Peptidoglycan recognition proteins: a novel family of four human innate immunity pattern recognition molecules. J Biol Chem. 2001;276(37):34686–34694.
- Werner T, et al. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc Natl Acad Sci U S A. 2000;97(25):13772–13777.
- Royet J, et al. Peptidoglycan recognition proteins: modulators of the microbiome and inflammation. Nat Rev Immunol. 2011;11(12):837–851.
- Node K, et al. Anti-inflammatory properties of cytochrome P450 epoxygenase-derived eicosanoids. Science. 1999;285(5431):1276–1279.
- Spector AA, Norris AW. Action of epoxyeicosatrienoic acids on cellular function. Am J Physiol Cell Physiol. 2007;292(3):C996–1012.
- Argiriadi MA, et al. Detoxification of environmental mutagens and carcinogens: structure, mechanism, and evolution of liver epoxide hydrolase. Proc Natl Acad Sci U S A. 1999;96(19):10637–10642.
- Grant DF, et al. Development of an in situ toxicity assay system using recombinant baculoviruses. Biochem Pharmacol. 1996;51(4):503–515.
- Borhan B, et al. Mechanism of soluble epoxide hydrolase. Formation of an alpha-hydroxy ester-enzyme intermediate through Asp-333. J Biol Chem. 1995;270(45):26923–26930.
- Sinal CJ, et al. Targeted disruption of soluble epoxide hydrolase reveals a role in blood pressure regulation. J Biol Chem. 2000;275(51):40504–40510.
- Deng Y, et al. Endothelial CYP epoxygenase overexpression and soluble epoxide hydrolase disruption attenuate acute vascular inflammatory responses in mice. FASEB J. 2011;25(2):703–713.
- Ulu A, et al. Anti-inflammatory effects of ω-3 polyunsaturated fatty acids and soluble epoxide hydrolase inhibitors in angiotensin-II-dependent hypertension. J Cardiovasc Pharmacol. 2013;62(3):285–297.
- Zhang W, et al. Soluble epoxide hydrolase gene deficiency or inhibition attenuates chronic active inflammatory bowel disease in IL-10(-/-) mice. Dig Dis Sci. 2012;57(10):2580–2591.
- Norwood S, et al. Epoxyeicosatrienoic acids and soluble epoxide hydrolase: potential therapeutic targets for inflammation and its induced carcinogenesis. Am J Transl Res. 2010;2(4):447–457.View this article via: PubMed Google Scholar
- Snyder GD, et al. Evidence for a membrane site of action for 14,15-EET on expression of aromatase in vascular smooth muscle. Am J Physiol Heart Circ Physiol. 2002;283(5):H1936–H1942.
- Oni-Orisan A, et al. Dual modulation of cyclooxygenase and CYP epoxygenase metabolism and acute vascular inflammation in mice. Prostaglandins Other Lipid Mediat. 2013;104–105:67–73.View this article via: PubMed Google Scholar
- Decker M, et al. EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides. J Lipid Res. 2012;53(10):2038–2045.
- Kundu S, et al. Metabolic products of soluble epoxide hydrolase are essential for monocyte chemotaxis to MCP-1 in vitro and in vivo. J Lipid Res. 2013;54(2):436–447.
- Panigrahy D, et al. Epoxyeicosanoids stimulate multiorgan metastasis and tumor dormancy escape in mice. J Clin Invest. 2012;122(1):178–191.
- Tomlinson G, et al. TLR-mediated inflammatory responses to Streptococcus pneumoniae are highly dependent on surface expression of bacterial lipoproteins. J Immunol. 2014;193(7):3736–3745.
- Rao KM. MAP kinase activation in macrophages. J Leukoc Biol. 2001;69(1):3–10.View this article via: PubMed Google Scholar
- Dong J, et al. GHIP in Streptococcus pneumoniae is involved in antibacterial resistance and elicits a strong innate immune response through TLR2 and JNK/p38MAPK. FEBS J. 2014;281(17):3803–3815.
- Guha M, Mackman N. LPS induction of gene expression in human monocytes. Cell Signal. 2001;13(2):85–94.
- Ridley AJ. Rho GTPases and cell migration. J Cell Sci. 2001;114(pt 15):2713–2722.View this article via: PubMed Google Scholar
- Cox D, et al. Requirements for both Rac1 and Cdc42 in membrane ruffling and phagocytosis in leukocytes. J Exp Med. 1997;186(9):1487–1494.
- Bystrom J, et al. Endogenous epoxygenases are modulators of monocyte/macrophage activity. PLoS One. 2011;6(10):e26591.
- Salgado-Pabon W, Schlievert PM. Models matter: the search for an effective Staphylococcus aureus vaccine. Nat Rev Microbiol. 2014;12(8):585–591.
- Zhang D, et al. Homocysteine upregulates soluble epoxide hydrolase in vascular endothelium in vitro and in vivo. Circ Res. 2012;110(6):808–817.
- Ai D, et al. Angiotensin II up-regulates soluble epoxide hydrolase in vascular endothelium in vitro and in vivo. Proc Natl Acad Sci U S A. 2007;104(21):9018–9023.
- Revermann M, et al. Soluble epoxide hydrolase deficiency attenuates neointima formation in the femoral cuff model of hyperlipidemic mice. Arterioscler Thromb Vasc Biol. 2010;30(5):909–914.
- Morisseau C, Hammock BD. Impact of soluble epoxide hydrolase and epoxyeicosanoids on human health. Annu Rev Pharmacol Toxicol. 2013;53:37–58.
- Theken KN, et al. Activation of the acute inflammatory response alters cytochrome P450 expression and eicosanoid metabolism. Drug Metab Dispos. 2011;39(1):22–29.
- Cheng PY, et al. Rapid transcriptional suppression of rat cytochrome P450 genes by endotoxin treatment and its inhibition by curcumin. J Pharmacol Exp Ther. 2003;307(3):1205–1212.
- Fisslthaler B, et al. EDHF: a cytochrome P450 metabolite in coronary arteries. Semin Perinatol. 2000;24(1):15–19.
- Kozak W, et al. 11,12-epoxyeicosatrienoic acid attenuates synthesis of prostaglandin E2 in rat monocytes stimulated with lipopolysaccharide. Exp Biol Med (Maywood). 2003;228(7):786–794.
- Chen BC, et al. Peptidoglycan induces nuclear factor-kappaB activation and cyclooxygenase-2 expression via Ras, Raf-1, and ERK in RAW 264.7 macrophages. J Biol Chem. 2004;279(20):20889–20897.
- Chen BC, et al. Rac1 regulates peptidoglycan-induced nuclear factor-kappaB activation and cyclooxygenase-2 expression in RAW 264.7 macrophages by activating the phosphatidylinositol 3-kinase/Akt pathway. Mol Immunol. 2009;46(6):1179–1188.
- Li P, et al. Epoxyeicosatrienoic acids enhance embryonic haematopoiesis and adult marrow engraftment. Nature. 2015;523(7561):468–471.
- Howe AK. Regulation of actin-based cell migration by cAMP/PKA. Biochim Biophys Acta. 2004;1692(2–3):159–174.View this article via: PubMed Google Scholar
- Dumaz N, Marais R. Integrating signals between cAMP and the RAS/RAF/MEK/ERK signalling pathways. Based on the anniversary prize of the Gesellschaft für Biochemie und Molekularbiologie Lecture delivered on 5 July 2003 at the Special FEBS Meeting in Brussels. FEBS J. 2005;272(14):3491–3504.
- Feng C, et al. Adenine nucleotides inhibit cytokine generation by human mast cells through a Gs-coupled receptor. J Immunol. 2004;173(12):7539–7547.
- Greenberg S. Modular components of phagocytosis. J Leukoc Biol. 1999;66(5):712–717.
- Beckett EL, et al. TLR2, but not TLR4, is required for effective host defence against Chlamydia respiratory tract infection in early life. PLoS One. 2012;7(6):e39460.
- Deng Y, et al. Cytochrome P450 epoxygenases, soluble epoxide hydrolase, and the regulation of cardiovascular inflammation. J Mol Cell Cardiol. 2010;48(2):331–341.
- Fleming I, Busse R. Endothelium-derived epoxyeicosatrienoic acids and vascular function. Hypertension. 2006;47(4):629–633.
- Haehnel V, et al. Transcriptional regulation of the human toll-like receptor 2 gene in monocytes and macrophages. J Immunol. 2002;168(11):5629–5637.
- Theken KN, Lee CR. Genetic variation in the cytochrome P450 epoxygenase pathway and cardiovascular disease risk. Pharmacogenomics. 2007;8(10):1369–1383.
- Yasar U, et al. Role of CYP2C9 polymorphism in losartan oxidation. Drug Metab Dispos. 2001;29(7):1051–1056.View this article via: PubMed Google Scholar
- Spiecker M, et al. Risk of coronary artery disease associated with polymorphism of the cytochrome P450 epoxygenase CYP2J2. Circulation. 2004;110(15):2132–2136.
- Morisseau C, et al. Effect of soluble epoxide hydrolase polymorphism on substrate and inhibitor selectivity and dimer formation. J Lipid Res. 2014;55(6):1131–1138.
- Nelson JW, et al. Soluble epoxide hydrolase dimerization is required for hydrolase activity. J Biol Chem. 2013;288(11):7697–7703.
- Edin ML, et al. Epoxide hydrolase 1 (EPHX1) hydrolyzes epoxyeicosanoids and impairs cardiac recovery after ischemia. J Biol Chem. 2018;293(9):3281–3292.
- Srivastava D. Microsomal epoxide hydrolase gene polymorphisms and susceptibility to prostate cancer: a systematic review. Indian J Cancer. 2016;53(2):213–215.
- Vaclavikova R, et al. Microsomal epoxide hydrolase 1 (EPHX1): Gene, structure, function, and role in human disease. Gene. 2015;571(1):1–8.
- Carey MA, et al. Pharmacologic inhibition of COX-1 and COX-2 in influenza A viral infection in mice. PLoS One. 2010;5(7):e11610.
- Zha W, et al. Functional characterization of cytochrome P450-derived epoxyeicosatrienoic acids in adipogenesis and obesity. J Lipid Res. 2014;55(10):2124–2136.
- Meerbrey KL, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci U S A. 2011;108(9):3665–3670.
- Vijayan D. Isolation and differentiation of monocytes-macrophages from human blood. Methods Mol Biol. 2012;844:183–187.View this article via: PubMed Google Scholar
- Mandell LA, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44(Suppl 2):S27–S72.
-
Version history
- Version 1 (September 30, 2021): In-Press Preview
- Version 2 (November 15, 2021): Electronic publication



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)











