Advertisement
Research ArticleImmunologyInfectious disease Free access | 10.1172/JCI96481
Clonally expanded γδ T cells protect against Staphylococcus aureus skin reinfection
Carly A. Dillen,1 Bret L. Pinsker,1 Alina I. Marusina,2 Alexander A. Merleev,2 Orly N. Farber,3 Haiyun Liu,1 Nathan K. Archer,1 Da B. Lee,1 Yu Wang,1 Roger V. Ortines,1 Steven K. Lee,1 Mark C. Marchitto,1 Shuting S. Cai,1 Alyssa G. Ashbaugh,1 Larissa S. May,4 Steven M. Holland,5 Alexandra F. Freeman,5 Loren G. Miller,6 Michael R. Yeaman,6,7,8,9 Scott I. Simon,10 Joshua D. Milner,3 Emanual Maverakis,2 and Lloyd S. Miller1,11,12,13
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Dillen, C. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Pinsker, B. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Marusina, A. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Merleev, A. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Farber, O. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Liu, H. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Archer, N. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Lee, D. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Wang, Y. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Ortines, R. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Lee, S. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Marchitto, M. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Cai, S. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Ashbaugh, A. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by May, L. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Holland, S. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Freeman, A. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Miller, L. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Yeaman, M. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Simon, S. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Milner, J. in: JCI | PubMed | Google Scholar
1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by
Maverakis, E.
in:
JCI
|
PubMed
|
Google Scholar
|

1Department of Dermatology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Department of Dermatology, School of Medicine, UCD, Sacramento, California, USA.
3Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
4Department of Emergency Medicine, School of Medicine, UCD, Sacramento, California, USA.
5Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA.
6Division of Infectious Diseases,
7Division of Molecular Medicine, and
8St. John’s Cardiovascular Research Center, Los Angeles Biomedical Research Institute, Harbor–UCLA Medical Center, Torrance, California, USA.
9Department of Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California, USA.
10Department of Biomedical Engineering, UCD, Davis, California, USA.
11Department of Medicine, Division of Infectious Diseases, and
12Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
13Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, Maryland, USA.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
Find articles by Miller, L. in: JCI | PubMed | Google Scholar
Published February 5, 2018 - More info
J Clin Invest. 2018;128(3):1026–1042. https://doi.org/10.1172/JCI96481.
Copyright © 2018, American Society for Clinical Investigation
Received: July 24, 2017; Accepted: December 19, 2017
-
Abstract
The mechanisms that mediate durable protection against Staphylococcus aureus skin reinfections are unclear, as recurrences are common despite high antibody titers and memory T cells. Here, we developed a mouse model of S. aureus skin reinfection to investigate protective memory responses. In contrast with WT mice, IL-1β–deficient mice exhibited poor neutrophil recruitment and bacterial clearance during primary infection that was rescued during secondary S. aureus challenge. The γδ T cells from skin-draining LNs utilized compensatory T cell–intrinsic TLR2/MyD88 signaling to mediate rescue by trafficking and producing TNF and IFN-γ, which restored neutrophil recruitment and promoted bacterial clearance. RNA-sequencing (RNA-seq) of the LNs revealed a clonotypic S. aureus–induced γδ T cell expansion with a complementarity-determining region 3 (CDR3) aa sequence identical to that of invariant Vγ5+ dendritic epidermal T cells. However, this T cell receptor γ (TRG) aa sequence of the dominant CDR3 sequence was generated from multiple gene rearrangements of TRGV5 and TRGV6, indicating clonotypic expansion. TNF- and IFN-γ–producing γδ T cells were also expanded in peripheral blood of IRAK4-deficient humans no longer predisposed to S. aureus skin infections. Thus, clonally expanded γδ T cells represent a mechanism for long-lasting immunity against recurrent S. aureus skin infections.
-
Introduction
Staphylococcus aureus is a Gram-positive bacterium that is the most common cause of skin infections in humans and is also an important cause of invasive and life-threatening infections, such as pneumonia, osteomyelitis, and bacteremia (1). The precise immune responses that protect against S. aureus skin infections are unclear, as nearly half of individuals with an S. aureus skin infection suffer a recurrence (2), despite the generation of high titers of specific antibodies and memory CD4+ T cells (3, 4). Moreover, all prior S. aureus vaccines in humans that targeted antibody-mediated phagocytosis have lacked efficacy or resulted in increased mortality (5). Therefore, a greater understanding of the adaptive immune responses that mediate long-lasting protection is needed to guide the future development of a successful S. aureus vaccine.
Neutrophils play an important role in host defense against S. aureus infections, as individuals with neutropenia (e.g., severe congenital neutropenia or as a result of chemotherapy) or impaired neutrophil function (e.g., chronic granulomatous disease) have a global susceptibility to S. aureus infections (6). However, certain primary immunodeficiency disorders have more selective impairment against S. aureus infections in the skin, including those in humans with defective IL-1R/TLR signaling (e.g., IRAK4 or MyD88 deficiency with impaired neutrophil recruitment; ref. 7–9) and in humans deficient in Th17 cells or IL-17 responses (i.e., autosomal dominant hyper-IgE syndrome and IL-17F or IL-17Ra deficiency; ref. 10–12). Similarly, MyD88-deficient mice have impaired neutrophil recruitment and host defense against S. aureus skin infections, predominantly due to loss of IL‑1β/IL‑1R1/MyD88 signaling (13, 14) and subsequent T cell–mediated IL-17 responses (15–18).
Nevertheless, the responses that mediate durable immunity against recurrent S. aureus skin infections likely involve mechanisms beyond MyD88/IRAK4 signaling and Th17/IL-17 responses because the childhood susceptibility to S. aureus skin infections in humans with MyD88 or IRAK4 deficiency wanes in adulthood (19) and humans with genetic defects in IL‑17 responses suffer from mucocutaneous candidiasis more commonly than S. aureus skin infections (20). Therefore, we set out to identify these concomitant protective immune responses that develop following a primary S. aureus skin infection that provide long-lasting protection against a secondary challenge.
-
Results
Protection of IL-1β–deficient mice against S. aureus skin reinfection. To assess whether immune protection developed following an S. aureus skin infection, WT C57BL/6 mice underwent a primary S. aureus skin infection (1°) in the lower back followed by a secondary S. aureus skin infection (2°) in a distant uninvolved site on the upper back on day 28 (d28) (Figure 1A). Both 1° and 2° WT mice developed skin lesion sizes (Figure 1, B and C) and bacterial burdens (measured by in vivo bioluminescence imaging and ex vivo CFU counting) (Figure 1, D–F) that did not significantly differ from each other, similarly to what occurred in prior reports (17, 18, 21). Given these results, we hypothesized that the normal activity of IL-1β in WT mice resulted in an effective response during both the 1° and 2° infections, making it difficult to observe an additional effect of any adaptive immune responses that developed. Therefore, we evaluated the 1° and 2° S. aureus skin infections in IL‑1β–/– mice, which have impaired neutrophil recruitment and host defense during a 1° S. aureus skin infection (13). The 1° IL-1β–/– mice developed markedly larger lesions and increased bacterial burden compared with WT mice (Figure 1, B–F). In contrast, 2° IL-1β–/– mice were protected and their responses were similar to those of WT mice. The protection was long term and not limited to a specific skin location, since 2° IL-1β–/– mice were still protected when the convalescent interval was increased to 8 or 20 weeks (Figure 1, G–J) or when the locations for 1° and 2° inoculations were reversed (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI96481DS1).
 Figure 1
Figure 1IL-1β–/– mice are protected against an S. aureus skin reinfection. (A) Time line for S. aureus skin reinfection model. (B) Representative photographs of skin lesions. (C) Mean total lesion size (cm2) ± SEM (n = 10/group). (D) Representative S. aureus in vivo bioluminescent signals. (E) Mean total flux (photons/s) ± SEM (n = 10/group). (F) Ex vivo CFUs from d7 infected skin (n = 5/group). (G–J) Mean total lesion size (cm2) ± SEM and mean total flux (photons/s) ± SEM after 8-week (G and H) or 20-week (I and J) convalescent period (n = 5–10/group). †P < 0.01,; ‡P < 0.001, compared with 1° mice, as calculated by 2-way ANOVA (C, E, G–J) or 2-tailed Student’s t test (F). Results in B–E and G and H are a compilation of 2 independent experiments. Results in F are representative of 2 independent experiments.
Neutrophil recruitment in reinfected IL-1β–deficient mice. By histology, 1° and 2° WT mice developed neutrophil abscesses at the peripheral edges and a dense band of Gram-positive S. aureus bacteria in the center (Figure 2, A–F). 1° IL-1β–/– mice had defective neutrophil abscess formation with an increased band length of Gram-positive bacteria. In contrast, 2° IL-1β–/– mice had restored neutrophil abscess formation with an increase in circulating neutrophils (Figure 2G). Neutrophils differentiate into distinct effector subsets after an S. aureus infection (22), suggesting they might mediate trained immunity against a subsequent S. aureus challenge as previously described (23). To evaluate this possibility, neutrophils from d28 IL-1β–/– mice were transferred to naive WT and IL-1β–/– mice 2 hours prior to 1° S. aureus skin infection. However, this did not result in any protection (Supplemental Figure 2, A and B).
 Figure 2
Figure 2Neutrophil recruitment is restored in IL-1β–/– mice during secondary infection. (A–D) Representative H&E- (A–C) and Gram-stained (D) histologic sections at 3 days after S. aureus inoculation. Scale bars: 240 μm. B and D show higher magnification of the black boxed area in A. C shows higher magnification of the white boxed area in B (n = 5/group). (E) Mean abscess area (cm2) ± SEM and (F) mean bacterial band width (mm) ± SEM from histologic sections (n = 5/group). (G) Mean neutrophil (PMN) number per 10 μl of blood ± SEM from 0 to 6 hours after S. aureus inoculation (n = 5/group). *P < 0.05; †P < 0.01, compared with 1° mice, as calculated by 2-tailed Student’s t test (E–G). Results in G are representative of 2 independent experiments.
Role of antibodies in conferring protection. Next, we evaluated whether more conventional adaptive immune responses contributed to the protection in 2° IL-1β–/– mice. d28 WT and IL-1β–/– mice had higher total IgG levels and S. aureus–specific IgG titers in the serum compared with naive mice (Figure 3A). There was also increased S. aureus–specific IgG in the infected skin of 2° WT and IL-1β–/– mice compared with naive mice (Figure 3B). To determine whether these antibodies conferred protection, serum from d28 IL-1β–/– mice was transferred to naive WT and IL-1β–/– mice prior to 1° S. aureus skin infection (Figure 3C). However, this passive transfer of S. aureus–specific antibodies did not confer protection in 1° WT or IL-1β–/– mice (Figure 3D).
 Figure 3
Figure 3Specific antibodies are not involved in protective immunity. (A) Mean total and S. aureus–specific serum IgG (pg/ml) ± SEM at d28 (n = 5/group). (B) Mean total and S. aureus–specific IgG levels IgG (pg/mg tissue weight) ± SEM in skin homogenates at 1 day after S. aureus inoculation (n = 5/group). (C) Time line of i.v. serum transfer. (D) Mean total lesion size (cm2) ± SEM and mean total flux (photon/s) ± SEM (n = 10/group). †P < 0.01; ‡P < 0.001, as measured by 2-tailed Student’s t test (A and B). Results in D are a compilation of 2 independent experiments.
Role of T cells in mediating protection. To determine whether the observed protection was mediated by lymphocytes either residing in the skin or trafficking from LNs, mice were treated with FTY720, which inhibits lymphocyte efflux from LNs (24). FTY720 treatment of 2° IL-1β–/– mice resulted in increased lesion sizes and bacterial burden (Figure 4, A and B) and loss of neutrophil abscess formation (Figure 4, C–G), similarly to what occurred in 1° IL-1β–/– mice. Thus, trafficking lymphocytes from draining LNs promoted the protective neutrophil recruitment response. This role for trafficking lymphocytes was only observed in 2° IL-1β–/– mice, as FTY720 treatment had no effect on 2° WT mice. LN cells harvested from d28 IL-1β–/– mice and transferred to naive IL-1β–/– mice prior to 1° S. aureus skin infection also conferred protection (Figure 4, H and I). Furthermore, the protective response was unique to LN cells from previously infected IL-1β–/– mice, since the transfer of LN cells from naive IL-1β–/– mice to naive IL-1β–/– mice had no effect (Supplemental Figure 3).
 Figure 4
Figure 4T cells trafficking from draining LNs mediate the protection. (A) Time line of FTY720 administration with (B) mean total lesion size (cm2) ± SEM and mean total flux (photon/s) ± SEM (n = 10/group). (C) Representative H&E-stained histologic sections at 3 days after S. aureus inoculation. Scale bars: 240 μm. D shows higher magnification of the black boxed area in C. E shows higher magnification of the white boxed area in D (n = 4/group). (F) Mean abscess area (cm2) ± SEM and (G) mean bacterial band width (mm) ± SEM from histologic sections (n = 4/group). (H) Draining LN cells harvested from d28 IL-1β–/– mice and transferred i.v. 1 day prior to 1° S. aureus inoculation of naive IL-1β–/– mice and (I) mean total lesion size (cm2) ± SEM and mean total flux (photon/s) ± SEM (n = 5/group). (J) Time line of anti-CD4 treatment and (K) mean total lesion size (cm2) ± SEM and mean total flux (photon/s) ± SEM (n = 5/group). †P < 0.01, compared with control 1° or 2° mice (B and I) as measured by 2-way ANOVA. Results in B, I, and K are a compilation of 2 independent experiments.
Since Th17 cells and Th1 cells have been implicated in host defense against S. aureus skin infections in humans (4, 10–12, 25–27) and mice (15–18), a role for CD4+ T cells was evaluated using anti-CD4 antibody depletion. Treatment successfully depleted CD4+ T cells from the blood and LNs (91% and 97%, respectively) while not significantly affecting the numbers of CD8+ T cells (Supplemental Figure 4). However, anti-CD4 antibody treatment had no effect on 2° IL-1β–/– or WT mice (Figure 4, J and K), indicating that CD4+ T cells were not responsible for the protection observed.
γδ T cells from LNs mediate protection. During the innate immune response that occurs within hours of an S. aureus skin infection in naive WT mice, IL-1β induces γδ T cells to promote IL-17–mediated neutrophil recruitment (16). However, whether γδ T cells contribute to host defense against a 2° S. aureus skin infection is unknown. To evaluate this possibility, total LN cells from d28 IL‑1β–/– mice were depleted of non–T cells; this was followed by isolating γδ T cells (TCRγδ+ cells) using magnetic bead separation. The enriched γδ T cells or the “flow-through” (CD3+ lymphocytes devoid of γδ T cells) (Supplemental Figure 5) was transferred to naive IL-1β–/– mice prior to 1° S. aureus skin infection (Figure 5A). Remarkably, transfer of only 50,000 γδ T cells mediated protection to a greater extent than 5 million other CD3+ T cells in the flow-through (Figure 5B). In addition, to evaluate for synergistic protection between γδ and CD4+ T cells as reported for other bacterial infections (28), lesion sizes and in vivo bioluminescence were determined in naive IL‑1β–/– mice that received no cell transfer or transfer of only γδ T cells (50,000 cells), only CD4+ T cells (5 million cells), or both γδ and CD4+ T cells combined (Figure 5, C and D). Transfer of only γδ T cells had the same protective effect as shown in Figure 5B. However, transfer of only CD4+ T cells or γδ and CD4+ T cells combined resulted in lesion sizes and in vivo bioluminescent signals that did not significantly differ from those of naive IL-1β–/– mice, indicating that CD4+ T cells did not have a protective effect either alone or when combined with γδ T cells.
 Figure 5
Figure 5γδ T cells from draining LNs confer protection. (A) Draining LNs were harvested from d28 IL-1β–/– mice, and γδ T cells or other CD3+ T cells were transferred i.v. 1 day prior to S. aureus inoculation of naive IL-1β–/– mice. (B) Total lesion size (cm2) ± SEM and mean total flux (photon/s) ± SEM (n = 10/group). (C) Draining LNs were harvested from d28 IL-1β–/– mice and γδ T cells, CD4+ T cells, or γδ T cells and CD4+ T cells combined were transferred i.v. 1 day prior to S. aureus inoculation of naive IL-1β–/– mice. (D) Total lesion size (cm2) ± SEM and mean total flux (photon/s) ± SEM (n = 5/group). *P < 0.05; †P < 0.01, between the 2 groups (B) or compared with IL-1β–/– mice without cell transfer (1° IL-1β–/–) (D) as measured by 2-way ANOVA. Results in B are a compilation of 2 independent experiments, and results in D are representative of 2 independent experiments.
Mechanisms that induce protective γδ T cells. Since protective γδ T cells developed in the absence of IL-1β activity, we hypothesized that an alternative MyD88 signal induced their generation and/or expansion. We first evaluated IL-1α because, like IL-1β, it signals via IL-1R1/MyD88. To inhibit both IL-1α and IL‑1β activity, IL-1β–/– mice were treated with an anti–IL-1R1 blocking antibody throughout the 42-day experiment (Figure 6, A and B). This treatment did not alter the 1° infection outcome nor did it diminish the protection observed in 2° IL-1β–/– mice, indicating that IL‑1α was not involved in the protection. Next, a role for TLR2, which recognizes S. aureus lipopeptides, lipoteichoic acid (LTA), and peptidoglycan (PGN) (6), was evaluated by treating TLR2–/– mice with an anti–IL-1R1 blocking antibody (Figure 6, C and D). Under these conditions, both TLR2 and IL-1R1 activity were blocked and this resulted in loss of protection during the 2° infection, indicating that TLR2 compensated for the lack of IL-1β to promote the protective response. Consistent for a role of TLR2 on γδ T cells, an appreciable percentage of γδ T cells in LNs of naive and d28 IL-1β–/– mice expressed TLR2, especially compared with the almost complete absence of TLR2 expression on CD4+ T cells (Figure 6, E and F). Furthermore, to evaluate for intrinsic TLR2/MyD88 signaling in T cells, 1° and 2° S. aureus skin infections were induced in Lck-cre×MyD88fl/fl mice, which have MyD88 specifically deleted in all T cell subsets, including γδ T cells (Figure 6, G and H). Lck-cre×MyD88fl/fl mice had a marked host defense impairment during the 1° infection and completely failed to develop any protective response during a 2° S. aureus skin infection, suggesting that T cell–intrinsic MyD88 signaling was required for mediating the protective response. Thus, TLR2 provided an alternative MyD88 signal in T cells that contributed to the protection observed in 2° IL-1β–/– mice.
 Figure 6
Figure 6γδ T cells are induced by T cell–intrinsic TLR2/MyD88 signaling. IL-1β–/– mice (A and B) or TLR2–/– mice (C and D) were treated with an α–IL-1R antibody or isotype control every other day beginning at d–1 and continuing throughout the 1° and 2° infection with total lesion size (cm2) ± SEM and mean total flux (photon/s) ± SEM (n = 5/group) determined. (E and F) Representative flow plots (E) and mean percentage ± SEM (F) of TLR2-expressing γδ and CD4+ T cells in inguinal LNs of naive and d28 IL‑1β–/– mice (n = 5/group). (G and H) Lck-MyD88–/– or WT mice (n = 5) during 1° and 2° infection with mean total lesion size (cm2) ± SEM and mean total flux (photon/s) ± SEM (n = 5/group) determined. *P < 0.05; †P < 0.01, compared with control 1° or 2° mice as measured by 2-way ANOVA. Results (A–H) are representative of 2 independent experiments.
TNF and IFN-γ, but not IL-17 or IL-22, mediate protection. To identify the effector cytokines produced by the protective γδ T cells, total LN cells from naive and d28 WT and IL-1β–/– mice were stimulated ex vivo with PMA/ionomycin and intracellular FACS was performed. IL-17A and IL-22 were first evaluated because they have been reported to be produced by trafficking γδ T cells (primarily in an IL-1β– and IL-23–dependent manner) during S. aureus skin infections (16), skin inflammation (24, 29–31), or after repeated intraperitoneal exposure to S. aureus (32). LN cells from d28 IL-1β–/– mice had either unchanged or decreased percentages of IL-17A+ and IL-22+ γδ T cells compared with naive IL‑1β–/– mice (Figure 7, A and B). Certain circulating γδ T cell subsets can produce TNF or IFN-γ (33–35); however, it is unknown whether TNF- and/or IFN-γ–producing γδ T cells developed after exposure to S. aureus and contributed to the protection observed. LN cells from d28 IL-1β–/– mice had significantly increased percentages of TNF+ and TNF+IFN-γ+ γδ T cells compared with naive IL‑1β–/– mice (Figure 7, A and B). In contrast, LN cells from d28 WT mice had an increased percentage of IL-17A+ γδ T cells and had either unchanged or decreased percentages of IL-22+, TNF+, and TNF+IFN-γ+ γδ T cells compared with naive WT mice.
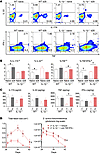 Figure 7
Figure 7γδ T cells produced TNF and IFN-γ to mediate protection. (A and B) Representative flow plots (A) and mean percentage ± SEM (B) are shown for IL‑17A–, IL-22–, TNF-, and IFN-γ–producing γδ T cells in inguinal LNs of naive and d28 WT and IL‑1β–/– mice (n = 5/group). (C) Protein levels of IL17A, IL-22, TNF, and IFN-γ in skin homogenates at d1 after S. aureus inoculation (n = 5/group). (D) Mean total lesion size (cm2) ± SEM and mean total flux (photon/s) ± SEM of IL-1β–/– mice ± anti-TNF/IFN-γ treatment (n = 5/group). *P < 0.05; †P < 0.01, as calculated by 2-tailed Student’s t test (B and C) or 2-way ANOVA (D). Results are representative of 2 independent experiments.
To evaluate whether these responses were relevant in vivo, these cytokine protein levels were measured in the S. aureus–infected skin of IL-1β–/– and WT mice (Figure 7C). In 2° IL-1β–/– mice, TNF and IFN-γ levels increased 5- and 2-fold, respectively, whereas IL-17A and IL-22 levels were not significantly different than in 1° IL-1β–/– mice. In contrast, IL-17A, IL-22, TNF, and IFN-γ levels were not significantly different between 1° and 2° WT mice. Finally, treatment of 2° IL-1β–/– mice with a combination of anti-TNF and anti–IFN-γ neutralizing antibodies resulted in loss of protection (Figure 7D), indicating that TNF and/or IFN-γ were crucial for mediating protection.
γδ T cells clonally expand in LNs following S. aureus skin infection. TCR complementarity-determining region 3–encoding (CDR3-encoding) sequences were mined from RNA-sequencing (RNA-seq) data sets of naive and d28 WT and IL-1β–/– mice for T cell repertoire analysis (Figure 8, A and B). The LNs of naive WT and IL-1β–/–mice had diverse αβ and γδ T cell repertoires. In LNs of uninfected mice, there were “public” (present in multiple mice) CDR3 aa sequences encoded by TRA (CVVGDRGSALGRLHF) and TRB (CASSLGGNYAEQFF) at low frequencies (~0.5% and ~0.1%, respectively), but these did not expand following S. aureus skin infection. In LNs of d28 WT and IL‑1β–/– mice, TRA and TRB repertoire analysis revealed no significant public S. aureus–induced expansions (Figure 8A). In contrast, LNs of d28 WT and IL-1β–/– mice exhibited strong γδ T cell expansions, as evidenced by the emergence of “top” (dominant) CDR3-encoding reads for TCR γ (TRG) (CACWDSSGFHKVF) and TRD (CGSDIGGSSWDTRQMFF), which represented approximately 20% of the total TRG and TRD CDR3-encoding sequences in the LNs following S. aureus skin infection (Figure 8B). The frequency of the TRG CACWDSSGFHKVF CDR3 aa sequence was increased in skin-draining LNs of 5 of 5 d28 WT mice and 6 of 7 d28 IL-1β–/– mice (Figure 8B) and was encoded by different TRG gene rearrangements, indicating that the expansion arose from multiple γδ T cells rather than a single clone. For example, while there were “canonical” TRG CACWDSSGFHKVF-encoding TCR gene rearrangements lacking N additions, this CDR3 aa sequence was also encoded for by gene rearrangements containing N additions (Table 1). Furthermore, this CDR3 aa sequence was encoded by 2 different 3′ V regions, TRGV5 and TRGV6 (Table 2), again indicating clonotypic T cell expansion. Curiously, a single d28 IL‑1β–/– mouse did not expand the TRGV5/6-encoded CACWDSSGFHKVF sequence in response to S. aureus, but instead utilized TRGV1 and TRGV3 gene segments to generate similar CDR3 aa sequences (i.e., CAVWTYSSGFHKVF and CAVWLYSSGFHKVF). The top TRD4 (CGSDIGGSSWDTRQMFF) CDR3 aa sequence also exhibited clonotypic expansion, as it was encoded by gene rearrangements with and without N additions in all d28 WT and IL‑1β–/– mice (Tables 3 and 4).
 Figure 8
Figure 8Clonotypic T cell expansion in response to S. aureus. CDR3 sequences were mined from RNA-seq data set of LNs and skin samples of naive WT and IL-1β–/– mice (n = 5/group) and d28 LNs (WT mice, n = 5; IL-1β–/– mice, n = 7). (A and B) Pooled results from all LN samples presented as the CDR3 aa sequence rank (x axis) versus proportion of the total CDR3 aa reads occupied by that particular CDR aa sequence (y axis). Blue dots indicate each of the different CDR3 aa reads; orange dots indicate public (found in most samples) TRA- and TRB-encoded CDR3 aa sequences; red dots indicate top (dominantly expanded) TRG- and TRD-encoded CDR3 aa sequences in d28 LNs of WT and IL‑1β-/- mice.
To verify these results, we mined the RNA-seq data set of Brady et al., which evaluated S. aureus–infected ear skin of WT mice on d0, d1, d4, and d7 (36). This revealed the same clonotypic TRG5/6-encoded CACWDSSGFHKVF sequence (Table 1), demonstrating that the clones detected in our experiments were also found in naive and S. aureus–infected WT skin in a separate mouse colony. We also performed CDR3-dedicated T cell repertoire analysis of skin samples obtained from WT and IL-1β–/– naive mice and d31 IL-1β–/– mice (3 days after the 2° S. aureus inoculation). In all cases, the TRGV5/6-encoded CACWDSSGFHKVF sequence and the TRDV4-encoded CGSDIGGSSWDTRQMFF sequence were detected (Tables 1 and 3), indicating that the same clonotypic γδ T cells were present in the skin both at baseline and during the 2° infection in WT and IL-1β–/– mice. Importantly, the TRGV5/V6-encoded CACWDSSGFHKVF and TRDV4-encoded CGSDIGGSSWDTRQMFF clonotypes were detected as unexpanded members of the skin-draining LNs of naive WT and IL-1β–/– mice (Figure 8B), indicating that these skin-resident γδ T cell clones were also low-frequency members of the peripheral T cell repertoire prior to infection.
The Spearman’s rank-order correlation coefficient was used to determine correlations between reads mapping to the TRGV5/V6-encoded CACWDSSGFHKVF sequence or the TRDV4-encoded CGSDIGGSSWDTRQMFF sequence and reads mapping to the T cell–associated transcription factors TBX21, RORC, GATA3, and FOXP3 (Table 5). The only significant positive correlations were between the 2 CDR3-encoding transcripts and TBX21, which encodes for the IFN-γ–inducing transcription factor T-bet, consistent with the increased percentage of IFN-γ+ γδ T cells in LNs of d28 IL-1β–/– mice (Figure 6, A and B). There were also significant negative correlations between the TRV5/TRV6 CDR3 transcripts and GATA3 (P < 0.05), which encodes for a Th2 cytokine–inducing transcription factor.
Finally, TRGV4-encoded CDR3 sequences detected in the skin were compared with those detected in the LNs before and after the S. aureus skin infection. These cells were singled out for additional analysis because prior reports found that resident and recruited dermal γδ T cells in response to inflammation or infection comprised IL-17–producing Vγ4+ γδ T cells (24, 29–31). Although the TRGV4 population was diverse, there were several public clones, the most common of which was a TRGV4/TRGJ1 clone whose CDR3 aa sequence was CSYGYSSGFHKVF. This clone was detected in 100% of the LN samples and 50% of the skin samples from naive IL-1β–/– and WT mice. However, it was not expanded in the d28 IL-1β–/– mice following S. aureus skin infection.
Humans with IRAK4 deficiency have a similar population of circulating γδ T cells. To ascertain whether similar TNF/IFN-γ circulating γδ T cells exist in humans, peripheral blood was evaluated from healthy individuals and individuals with primary immunodeficiency disorders with an increased susceptibility to S. aureus skin infections and other pyogenic infections, including IRAK4 deficiency with impaired neutrophil migration due to defective IL‑1R/TLR signaling (including a lack of IL-1β activity, as in IL-1β–/– mice) (7, 8) and chronic granulomatous disease (CGD) with defective neutrophil killing due to loss-of-function mutations in NADPH oxidase (37). Healthy and IRAK4-deficient individuals had 1%–5% γδ T cells, whereas individuals with CGD had only approximately 1% γδ T cells of the total CD3+ T cell population (Figure 9, A and B). Healthy individuals had virtually equivalent percentages (~45%) of Vδ1+ and Vδ2+ γδ T cells (Figure 9, C and D), which are the major γδ T cell populations in human blood (33–35). γδ T cells were mostly Vδ2+ (>80%) in IRAK4 deficiency, and in contrast, the few γδ T cells in CGD were mostly Vδ1+ (~80%). After PMA/ionomycin stimulation and intracellular FACS analysis, the percentages of TNF- and/or IFN-γ–producing cells were evenly distributed (20%–35%) among Vδ2+ and Vδ1+ cells in healthy individuals. In contrast, almost all of TNF– and/or IFN-γ–producing cells (88%–96%) were Vδ2+ cells in IRAK4 individuals and a broad range of TNF– and/or IFN-γ–producing cells (38%–92%) were Vδ1+ cells in CGD individuals (Figure 9, E–H). The cellular phenotype of blood drawn from the IRAK4 patient at the end of and several years after her recurrent infections was very stable. Interestingly, the percentage of γδ T cells that produced IL-17A among all human experimental groups was less than 0.68%, which was 50- to 100-fold lower than the percentage of TNF and/or IFN-γ–producing γδ T cells (Figure 9, I and J), indicating that IL‑17A is not a major cytokine produced by circulating γδ T cells in humans.
 Figure 9
Figure 9TNF/IFN-γ–producing γδ T cells from PBMCs in individuals with IRAK4 deficiency and CGD. PBMCs from healthy controls (ctrl, n = 6), patients with IRAK4 deficiency (n = 3 samples, including 2 samples from the same individual at age 17, toward the end of the predilection for recurrent infection, and at age 22), and patients with CGD (n = 4) were analyzed. Representative flow plots and mean percentage ± SEM are shown for total γδ T cells in PBMCs (A and B), Vδ1+ versus Vδ2+ γδ T cells (C and D), TNF production (E and F), IFN-γ production (G and H), and IL-17A production (I and J). *P < 0.05; †P < 0.01, as measured by 2-tailed Student’s t test with Bonferroni’s correction.
-
Discussion
The immune mechanisms that mediate long-lasting protection to S. aureus skin infections have remained elusive, especially since recurrences are common despite the generation of high titers of specific antibodies and memory αβ T cells (3, 4). In the present study, we utilized S. aureus–susceptible IL-1β–/– mice with impaired neutrophil recruitment and IL-17 responses to elucidate host defense mechanisms that provide durable protection against an S. aureus skin reinfection. We report that, following an initial S. aureus skin infection, clonotypic TNF/IFN-γ–producing γδ T cells expanded in skin-draining LNs and protected against a subsequent S. aureus skin challenge, which likely complements the previously characterized antibody and IL-17 responses in immunity to S. aureus skin infections (15–18). This γδ T cell protective response was long lived, as it was still present at 20 weeks, similar to αβ T cell memory responses in other models (38, 39). The protection was specific to γδ T cells, since transfer of γδ T cells, but not serum containing S. aureus–specific antibodies, CD4+ T cells, or primed neutrophils from previously infected IL‑1β–/– mice, rescued the immune impairment of naive IL-1β–/– mice.
In mice, γδ T cells have been increasingly recognized as contributing to both innate and adaptive immune responses (33–35). During development, murine γδ T cells leave the thymus, possess invariant TCRs, and populate various epithelial sites in tissues and organs (33–35), such as the Vγ5+ γδ T cells in mouse epidermis (40). In general, circulating murine γδ T cells produce IL-17 or IFN-γ, with IL‑1β/IL-1R1 activation promoting IL-17 production (16, 29, 32, 41). Since data in humans indicate that protective immunity to recurrent S. aureus skin infections can develop in the absence of MyD88/IRAK4 signaling (including IL‑1β/IL-1R1/MyD88 signaling) (19) and Th17/IL‑17 responses (20), we evaluated IL-1β–deficient mice to uncover previously unknown components of the S. aureus–directed immune response.
We found an important role for TNF and IFN-γ in protecting against a secondary S. aureus skin challenge, which was unexpected because TNF-αRI–/– and IFN-γR–/– mice were found to have no immune impairment against a 1° S. aureus skin infection (14, 16). However, a recent report found that deficiency in both IFN-γ and IL-17 signaling, but not either alone, resulted in spontaneous and persistent mucocutaneous S. aureus infections in mice (42), suggesting that both IFN-γ and IL-17 are required for durable protection against S. aureus skin infections. Furthermore, impaired Th1 responses (i.e., IFN-γ+ CD4+ T cells), but not IL-17/Th17 cell responses, were observed in the blood and S. aureus–infected skin of HIV+ individuals (known to be highly susceptible to S. aureus skin infections), implicating IFN-γ rather than IL‑17 in mediating protective immunity to S. aureus skin infections in humans (26). These data in mice and humans support our finding that TNF and IFN-γ responses are essential for long-lasting immunity against S. aureus skin infections.
In this study, TLR2 compensated for the lack of IL-1β, since the protection observed in the 2° IL-1β–/– mice was lost when both IL-1R1 and TLR2 were inhibited in vivo. In addition, TLR2 was expressed by γδ T cells in the LNs of IL-1β–deficient mice, and the effect of TLR2 was intrinsic to MyD88 signaling on T cells, as Lck-cre×MyD88fl/fl mice did not develop protection against 2° S. aureus skin infection. TLR2 was likely activated by known S. aureus–derived ligands for TLR2 (e.g., lipoproteins/lipopeptides, LTA, and PGN; ref. 6) in the infected skin and/or in the draining LNs to generate the protective γδ T cells that produced TNF/IFN-γ rather than IL‑17A. This is consistent with the lack of expansion of IL‑17A–producing γδ T cells in IL-1β–/– mice after the 1° S. aureus skin infection in vivo despite active TLR2/MyD88 signaling in these mice. Although a prior report found that TLR2 activation of γδ T cells in vitro resulted in IL-17A production, IL-1β was likely also produced in the cultures, providing an alternative explanation for the increased IL-17A production (41).
Regarding the specific CDR3 gene rearrangements, our results greatly modify the currently accepted views on γδ T cells. Early seminal discoveries demonstrated that the first murine γδ T cells produced by the thymus during early development have canonical TCRs lacking N additions, including an invariant δ chain (43, 44). It is believed that these T cells then migrate to distinctive sites within the body dictated by the TCR-γ chain they express (33, 45). The TRGV5 gene rearrangement detected in our study corresponds to that reported for the previously described invariant Vγ5+ dendritic epidermal T cells (DETCs) (43, 44). DETCs are the most common skin-resident γδ T cell population and have been reported to recognize a stress-induced self-antigen derived from keratinocytes (46, 47). In contrast, the TRGV6 gene rearrangement described herein corresponds to that reported for canonical γδ T cells residing in the liver, placenta, kidney, uterus, tongue and other mucosal sites (33, 45). Interestingly, the dominant TRDV4 gene rearrangement that was found in both the skin and LNs encodes the same CDR3 aa sequence for the Vδ1 chain of DETCs and was previously reported to only be present in remodeling epithelial tissues such as the placenta and lactating mammary glands (48, 49). Although still the dominant view, some reports have been inconsistent with the distribution of γδ T cells just described. For example, a recent report failed to identify invariant Vγ5+ γδ T cells in the mouse skin, but rather described an IL-17–producing CD3hi invariant Vγ6+ γδ T cell population that was increased following imiquimod-induced skin inflammation (50). Vγ6+ γδ T cells have also been shown to strongly produce IL-17 in response to fungal and bacterial infections, including repeated intraperitoneal exposure to S. aureus, at different anatomical sites (32, 51–53).
Our study differs substantially from these prior reports in that we used deep sequencing to characterize the γδ T cell repertoire. This revealed that invariant Vγ5+ and Vγ6+ T cells reside in the skin and in the skin-draining LNs. Furthermore, we demonstrate that both the classical TRGV5 and TRGV6 gene rearrangements encode for the exact same CDR3 aa sequence, CACWDSSGFHKVF, which we found is also encoded by noncanonical TRG gene rearrangements containing N additions. Finally, we demonstrate that following S. aureus skin infection, there is a clonotypic expansion of these TRGV5/V6 γδ T cells in the skin-draining LNs. Multiple gene rearrangements encoding for the same CDR3 aa sequence, as reported here, favor antigen specificity as the driving force behind the observed pathogen-induced T cell expansion. Although it is unclear whether the γδ T cell TCR reactivity was against a yet-to-be-described S. aureus–derived antigen or self-antigen as previously postulated (46, 47), it is clear that this distinctive TRGV5/V6-encoded CDR3 aa sequence is critical for mediating robust immune protection against pathogens at the skin and potentially other barrier sites. Furthermore, the TRGV5 and TRGV6 CDR3-encoding sequences correlated with TBX21 expression, providing an explanation for increased γδ T cell production of TNF/IFN-γ. The TRGV5/V6 response is in contrast to that observed for TRGV4 γδ T cells, which did not expand in response to S. aureus infection. TRGV4 γδ T cells were also found at a much lower frequency than the clonotypic TRGV5/V6 γδ T cells.
In humans, Vδ2+ γδ T cells have a semi-invariant TCR repertoire and rapidly expand and contribute to host defense against microbial infections (54, 55). Vδ1+ γδ T cells have a more diverse TCR repertoire and have been associated with adaptive responses, especially against viruses such as CMV (54, 55). In the present study, an increased proportion of circulating TNF/IFN-γ–producing Vδ2+ γδ T cells were found in IRAK4-deficient individuals; these most likely expanded as a result of the recurrent infections in these patients (8). In the setting of IRAK4 deficiency, the Vδ2+ γδ T cells might restore the impaired neutrophil recruitment response (7), which would explain the patients’ decreased susceptibility to S. aureus with age (8, 19). In contrast, Vδ2+ γδ T cells were virtually absent in individuals with CGD, a disease in which patients never overcome their predisposition to S. aureus and other pyogenic infections (37). In addition, almost all of the circulating Vδ1+ and Vδ2+ γδ T cells from all healthy humans and IRAK4 and CGD patients lacked the ability to produce IL-17 (or IL-22). This is in contrast to the high numbers of circulating IL-17–producing T cells in mice (56), suggesting that IL-17 responses might not be as relevant for circulating γδ T cells in humans. Moreover, since human circulating γδ T cells primarily produce TNF/IFN-γ rather than IL-17 or IL-22, the protective TNF/IFN-γ–producing γδ T cells that we identified in mice might better translate to protective γδ T cell responses in humans.
There are several limitations and future directions. First, a key question is whether the expanded γδ T cells from previously infected IL-1β–/– mice could confer protection to WT mice. To evaluate this, we performed an LN transfer from d28 IL-1β–/– mice to naive WT mice as in Figure 4H. However, this did not result in further protection of the WT mice (see Supplemental Figure 6). The lack of protection could have been due to normal IL‑1β activity in WT mice, which induced effective neutrophil recruitment that could not be further enhanced. Alternatively, the presence of IL‑1β activity could have negatively affected TNF and IFN-γ production by the transferred γδ T cells from IL‑1β–/– mice. A future direction will be to determine the mechanisms by which clonally expanded γδ T cells confer protection to WT mice. Second, transfer of CD4+ T cells to naive IL‑1β–/– mice did not have a protective effect. Further, transferring γδ and CD4+ T cells in combination resulted in loss of the protective effect of the γδ T cells. Although the reason for this is unclear, this result is consistent with recent findings that naturally generated or vaccine-induced CD4+ T cell responses were either not protective or resulted in increased mortality in mouse models of S. aureus bacteremia (57, 58). Third, the FTY720 experiments indicate that trafficking of the protective γδ T cells from LNs during the infection was required for protection, but it is unclear whether these cells might also function as tissue-resident cells. Regarding expression of homing and adhesion molecules of tissue-resident memory T cells, the γδ T cells from LNs of d28 IL-1β–/– mice had substantially decreased expression of CCR4 and significant but subtle differences in expression of CD103 and CLA compared with that in naive IL‑1β–/– mice (Supplemental Figure 7). Finally, it is unclear whether the TCR signaling in the γδ T cells was required for protection; however, the butyrophilin family members (e.g., Skint1, Skint3, or Skint9) have recently been implicated as TCR ligands or costimulatory molecules for DETCs, which express the same CDR3 aa sequence (59, 60). There was a significant correlation between reads encoding for the top γδ CDR3 sequences and BTLN2 (but not SKINT1, SKINT3, or SKINT9) in the LNs of d28 IL-1β mice (Supplemental Figure 8), suggesting that Btnl2 might activate the protective γδ T cells in our model. A role of TCR signaling and butyrophilin family members will be the subject of our future work.
Taken together, this study identified a clonotypic expansion of TNF/IFN-γ–producing γδ T cells that were induced by TLR2/MyD88 signaling to promote protection against an S. aureus skin reinfection. These findings provide important mechanistic insights for targeting specific γδ T cells in future vaccines and immunotherapies to promote durable immunity against S. aureus skin infections.
-
Methods
Staphylococcus aureus strain. The bioluminescent USA300 LAC::lux strain used in all experiments was previously generated from the community-acquired methicillin-resistant S. aureus (MRSA) USA300 LAC isolate obtained from a skin infection outbreak in the Los Angeles County Jail (Los Angeles, California, USA) and was provided by Tammy Kielian (University of Nebraska, Lincoln, Nebraska, USA). USA300 LAC::lux possesses a modified luxABCDE operon stably integrated in the bacterial chromosome so that the emission of blue-green light from live and metabolically active bacteria is maintained in all progeny without selection.
Bacterial preparation. S. aureus bacteria was streaked onto a tryptic soy agar (TSA) plate (tryptic soy broth [TSB] plus 1.5% bacto agar; BD Biosciences) and grown overnight at 37°C in a bacterial incubator. Single colonies were cultured in TSB at 37°C in a shaking incubator (240 rpm) overnight (18 hours), followed by a 1:50 subculture at 37°C for 2 hours to obtain midlogarithmic phase bacteria. The bacteria were pelleted, resuspended, and washed in PBS. The absorbance (A600) was measured to estimate the number of CFUs, which was verified after overnight culture on TSA plates.
Mice. C57BL/6 WT mice, TLR2–/– mice (B6.129-Tlr2tm1Kir/J), Lck-cre mice (B6.Cg-Tg(Lck-cre)548Jxm/J), and MyD88fl/fl (B6.129P2(SJL)-MyD88tm1Defr/J) mice were obtained from Jackson Laboratories. Lck-cre mice were crossed with MyD88fl/fl mice to obtain the Lck-cre × MyD88fl/fl mouse strain. IL-1β–/– mice on a C57BL/6 background were provided by Yoichiro Iwakura (University of Tokyo, Tokyo, Japan).
Mouse model of S. aureus skin reinfection. For each experiment, sex- and age-matched mice were used beginning at 6 and 8 weeks of age. For the 1° S. aureus skin infection, the dorsal backs of anesthetized mice (2% isoflurane) were shaved and injected intradermally (i.d.) in the lower back with 3 × 107 CFU of USA300 LAC::lux in 100 μl of PBS using a 29-gauge insulin syringe. After a 2-week convalescent period, on d28, a 2° S. aureus skin infection was performed by i.d. injection at a distant skin site in the upper back with the same inoculum and procedures as used for the 1° S. aureus skin infection. In some experiments, the convalescent period was increased to 8 or 20 weeks prior to the 2° S. aureus skin infection or the i.d. inoculation sites on the lower and upper backs of mice were reversed.
Measurement of total lesion area. Total lesion size (cm2) was measured from digital photographs of the skin of anesthetized mice (2% isoflurane) using ImageJ software (NIH) and a millimeter ruler as a reference.
In vivo bioluminescent imaging. In vivo bioluminescent imaging was performed on anesthetized mice (2% isoflurane) using a Lumina III IVIS (PerkinElmer) and total flux (photons/s) was measured within a 1 × 103 pixel circular region of interest using Living Image software (PerkinElmer) (limit of detection: 2 × 104 photons/s).
Skin tissue homogenates and bacterial quantification. Skin tissue homogenates were obtained by performing a 10-millimeter lesional skin punch biopsy (Acuderm) and homogenizing each specimen (Pro200 Series homogenizer; Pro Scientific) in Reporter Lysis buffer (Promega) containing protease inhibitor cocktail tablets (Roche Life Sciences) at 4°C. Ex vivo CFUs were counted after plating serially diluted skin tissue homogenates overnight on TSA plates.
Histology. Skin punch biopsy specimens (10 mm) were collected, fixed in formalin (10%), and embedded in paraffin. Sections (4 μm) were mounted onto glass slides and stained with H&E and Gram stain by the Johns Hopkins Reference Histology Laboratory.
Measurement of total and S. aureus–specific IgG. Levels of total IgG in serum and skin tissue homogenates were determined using a mouse IgG ELISA according to the manufacturer’s protocol (eBioscience). S. aureus–specific IgG levels in the specimens were obtained by coating ELISA plates overnight with heat-killed USA300 LAC::lux in the stationary phase and then proceeding with manufacturer’s instructions (eBioscience).
Serum isolation and transfer to mice. Mouse serum was obtained from newly collected blood (from terminal cardiac puncture on anesthetized mice), which was allowed to clot at room temperature for 30 minutes. Serum supernatants were then collected after centrifuging the clotted blood at 1,500 g for 10 minutes at 4°C. Serum samples were stored at –20°C prior to use. Serum (200 μl, 37°C) was transferred to mice via i.v. injection (via retroorbital vein) on day –1 (d–1) and d0 prior to S. aureus skin inoculation; serum used was 4 times that transferred in a prior study (18).
In vivo antibody and FTY administration. For CD4+ T cell depletion, mice were treated i.p. with anti-CD4 antibody (clone GK1.5, BioXCell) 300 μg in 500 μl PBS on d–1 and 100 μg in 500 μl sterile PBS on d0 and d7 of the 2° S. aureus skin infection, according to previously described dosing (18). Depletion of CD4+ T cells was confirmed from blood samples (collected via retroorbital venipuncture) immediately prior to the S. aureus skin infection by flow cytometry using antibodies against CD4 (clone RM4-5), CD8α (clone 53-6.7), and CD3ε (clone 145-2C11). For combined TNF and IFN-γ neutralization, mice were treated i.p. with anti-mouse TNF (clone XT3.11) and anti-mouse IFN-γ (clone XMG 1.2), with 200 μg of each given together in 500 μl sterile PBS on d–1, d0, and d1 of the 2° S. aureus skin infection, according to the manufacturer’s recommended dosing (BioXCell). To block IL-1R1, mice were treated i.p. with anti–IL-1R1 antibody (clone JAMA-147) 100 μg in 500 μl sterile PBS beginning on d–1, d0, and d1 of the 1° S. aureus skin infection and then every other day throughout the entire 42-day experiment (including during the 2° S. aureus skin infection), according to the manufacturer’s recommended dosing (BioXCell). All control mice were treated i.p. with a corresponding isotype control antibody (BioXCell) at the equivalent dose and volume. To block lymphocyte efflux from LNs, mice were treated i.p. with FTY720 (Sigma-Aldrich), 1 mg/kg in 100 μl sterile water, beginning on d27, d28, and d29 of the 2° S. aureus skin infection and continuing every other day as previously described (24).
Lymphocyte isolation, magnetic bead separation, and transfer to mice. A single suspension of LN cells was obtained after manually pushing draining LNs through a cell-separation filter (40 μm). Cells were counted and resuspended in MACS Separation Buffer (degassed PBS containing BSA [0.5%] and EDTA [2 mM]) (Miltenyi Biotec). γδ T cells were isolated using a TCRγ/δ+ Isolation Kit for mice, according to the manufacturer’s instructions (Miltenyi Biotec). Briefly, non–T cells were depleted using direct magnetic labeling of CD45R and CD11b microbeads. Next, γδ T cells were indirectly labeled using anti-TCRγ/δ-biotin, and anti-biotin microbeads were then used to positively select γδ T cells. The remaining T cells were collected via flow-through on the column. 50,000 γδ T cells or 5 million CD3+ T cells (devoid of γδ T cells) in the flow-through were transferred to mice i.v. (via retroorbital vein) at 1 day prior to S. aureus skin inoculation. The adoptively transferred cells were either 96.1% CD3+ T cells with 0.8% GL3+ γδ T cells collected from the flow-through or 95.7% GL3+ γδ T cells isolated from the column (Supplemental Figure 5). In a separate experiment, γδ T cells were isolated as described above, while the non–T cell–depleted fraction left over after this isolation was subsequently used to isolate CD4+ T cells using a CD4+ T cell isolation kit (Miltenyi Biotech) per the manufacturer’s instructions. One day prior to inoculation with S. aureus, 50,000 γδ T cells, 5 million CD4+ T cells, or 50,000 γδ T cells plus 5 million CD4+ T cells were injected via retroorbital vein. The purity percentages of isolated cells that were adoptively transferred were 95.7% GL3+ γδ T cells and 98.1% CD4+ T cells (Supplemental Figure 5).
Mouse cell phenotypic analysis. LNs were harvested from mice, and single-cell suspensions were created. Cells were resuspended in PBS containing 1% BSA. 1 × 106 Cells were stained for surface markers using antibodies against CCR4 (clone 2G12), CD4 (clone GK1.5), CD8α (clone 53-6.7), CD3 (clone REA641), CLA (HECA-452), TCRγδ (clone GL3), CD103 (clone REA789), and TLR2 (clone 6C2) for 30 minutes at 4°C. Cells were washed twice in PBS and resuspended in PBS with 1% BSA for acquisition. Propidium iodide was added at 1:100 immediately prior to acquisition for discrimination of dead cells. Intracellular staining was performed by first creating single-cell suspensions from LNs, which were resuspended in RPMI containing FBS (10%), penicillin (100 U/ml), and streptomycin (100 μg/ml). 1 × 106 Cells per well were plated in 96-well cell-culture plates in the presence of a cell-stimulation cocktail plus protein transport inhibitors (eBioscience) containing PMA, ionomycin, brefeldin A, and monensin. Control wells contained only protein transport inhibitor cocktail (brefeldin A and monensin). Cells were incubated at 37°C for 6 hours. Cells were washed once before staining for viability (Viability Fixable Dye, Miltenyi Biotec) and surface markers using antibodies against CD4 (clone RM4-5), CD8α (clone 53-6.7), and TCRγδ (clone GL3) for 30 minutes at 4°C. Cells were washed before being fixed using FACSFix (BD Biosciences) for 30 minutes. Cells were permeabilized by washing 3 times and then incubating for 10 minutes in Perm/Wash (BD Biosciences) and then incubated with antibodies against IL-17A (clone TC11-18H10), IL-22 (clone IL22JOP), TNF (clone MP6-XT22), and IFN-γ (clone XMG1.2) or isotype control mAbs (Miltenyi Biotec) (Supplemental Figure 9) for 30 minutes before being washed and resuspended. Cell acquisition was performed on a MACSQuant flow cytometer (Miltenyi Biotec), and data were analyzed using FlowJo software (Tree Star).
Cytokine protein levels. Protein levels of IL-22, IFN-γ, and TNF were measured from homogenized punch biopsy specimens collected at d1 after the S. aureus skin inoculation by using Bio-Plex protein assays and normalized to total protein according to the manufacturer’s recommendations (Bio-Rad). Protein levels of IL-17A were measured using an ELISA kit according to the manufacturer’s recommendations (R&D Systems). Data are presented as pg/ml (Bio-Plex) or pg/mg tissue weight (ELISA).
RNA isolation. Mouse LNs and skin biopsies were stabilized by addition of RNAlater (Ambion). Homogenization was performed using a TissueLyzer II (QIAGEN) at 20 Hz for 1 minute at –80 C. Total RNA was extracted using the RNeasy Plus Mini Kit (QIAGEN). RNA concentrations were then quantified using a Qubit Fluorometer, and RNA integrity was assessed using the Agilent TapeStation (Agilent). Samples with RNA integrity number (RIN) of 8 or more were used for this study.
RNA-seq. Indexed libraries were constructed from 1,000 ng of total RNA using the TruSeq Stranded mRNA Sample Prep Kit (Illumina) following the manufacturer’s instruction. The quantity and quality of the libraries were also assessed by Qubit and Agilent 2100 Bioanalyzer, respectively. The average library size was 400 bp. Library molar concentration was validated by quantitative PCR (qPCR) for library pooling. Sequencing was performed on the Illumina HiSeq 4000 platform using PE150 chemistry (Illumina). RNA-seq data were deposited in the NCBI’s Sequence Read Archive database (SRA SRP126124).
Analysis of public data sets. RNA-seq FASTQ files of mouse skin were downloaded from the NCBI SRA database (SRA SRP040121), and information about the samples is in the corresponding publication (36).
RNA-seq data analysis. Raw sequencing data were received in FASTQ format. Sequencing reads were mapped to the UCSC mm10 mouse reference genome by STAR. Gene-expression level normalization and differential expression analysis were carried out by using the DESeq2 Bioconductor R package. This package provides statistics for determination of differential expression using a model based on the negative binomial distribution. Genes with an adjusted P value of less than 0.05 and fold change greater than 2 were assigned as differentially expressed. In order to examine the relationship of gene expressions across the samples, Spearman’s correlation of normalized gene counts was computed. Spearman’s rank correlation coefficients and P values were calculated in R (version 3.1.2, http://www.R-project.org/) using the cor.test function.
Extraction of TCR sequence information from RNA-seq data. MiXCR software was used to extract TCR CDR3 sequences from RNA-seq data. Analyses were performed with the “-p rna-seq” option recommended for analysis of RNA-seq data.
CDR3 definition. The TCR CDR3 region was defined as the amino acid residues starting with the C at position 104 and ending with the F at position 118 based on International ImMunoGeneTics (IMGT) nomenclature and a TCR numbering system. Likewise, gene names of V and J regions are designated according to the IMGT name nomenclature for T cell receptors of mice.
Amplification of TCR CDR3. For targeted CDR3 sequencing, we synthesized cDNA with anchor sequence incorporation. This method is based on the 5′ template-switching ability of certain reverse transcriptases, which has been used for αβ T cell repertoire analysis (61). Reverse transcriptions were performed using the Ribo_TSO (5′-AAGCAGTGGTATCAACGCAGAGTACTCTT(rG)5-3′), TCR Cγ- and Cδ-specific primers mDC1R RT (5′-CACCAGACAAGCAACATTTG-3′), mGC1R RT (5′-CTTTTCTTTCCAATACACCCTTAT-3′), and SMARTScribe reverse transcriptases (Clontech) according to the manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) was performed for each sample to identify the minimum number of PCR cycles needed to ideally amplify TCR without overamplification. We carried out qRT-PCR iProof High-Fidelity Master Mix (Bio-Rad) and EvaGreen Dye (Biotium) with the following primer pairs: Smart20 (5′-CACTCTATCCGACAAGCAGTGGTATCAACGCAG-3′), mDC2R (5′-CATGATGAAAACAGATGGTTTGG-3′) for TCRδ, Smart20 (5′-CACTCTATCCGACAAGCAGTGGTATCAACGCAG-3′), and mGC2R (5′-GGAGATTTGTTTCAGCA-3′) for TCR-γ. The PCR cycle was as follows: 98°C for 2 minutes, 95°C for 20 seconds, 68°C for 20 seconds, and 72°C for 50 seconds for 40 cycles, followed by a final extension step of 72°C for 5 minutes. Once the optimal number of PCR cycles was established, final PCR was carried without EvaGreen. All amplification reactions were assayed in at least 10 separate tubes to minimize PCR amplification bias. The reaction products were purified using the MinElute PCR Purification Kit using Qiacube (QIAGEN) and quantified using a Qubit Fluorometer and Quant-iT dsDNA BR Assay Kit (Invitrogen).
TCR library preparation. All libraries from purified TCR amplicons were prepared using the Nextera XT Index Kit (Illumina Inc.) according to the manufacturer’s instructions for the 16S Metagenomic Sequencing Library Preparation protocol, with minor modifications. Briefly, first-stage PCR was performed with KAPA HiFi HotStart ReadyMix (KAPA Biosystems) and the primer pairs were designed according to protocol guidelines. Primer pairs were as follows: first-stage PCR forward primer: Nextera-Step_1: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCACTCTATCCGACAAGCAGT-3′; first-stage PCR reverse primer for TCRδ NextMusDCj: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTTTGGCCGGAGGCTGGCTTT-3′; and first-stage PCR reverse primer for TCR-γ NextMusDG2R: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGAGATTTGTTTCAGCA-3′. First-stage PCRs were purified using AMPure XP beads (Beckman Coulter Inc).
Illumina sequencing adapters and dual index barcodes were added in subsequent second-stage PCR using the Nextera XT indices and HotStart ReadyMix (KAPA Biosystems).
Indexed libraries were purified using 2 rounds of 0.7 volumes of AMPure XP beads (Beckman Coulter Inc.) and quantified using a Qubit Fluorometer and Quant-iT HS DNA Assay Kit (Invitrogen). The quality of the libraries was also assessed by Agilent 2100 Bioanalyzer, and the average library size was 600–700 bp.
TCR library sequencing and analysis. Libraries were pooled to a final pool concentration of 4 nM including a 10% PhiX Control v3 (Illumina) spike-in. Sequencing was performed on an Illumina MiSeq sequencer using the 600-cycle MiSeq Reagent Kit v3 (Illumina) with paired-end reads. Raw-sequencing reads were processed for FASTQ conversion and demultiplexing using the MiSeq Reporter. MiXCR software was used to extract TCR CDR3 sequences from sequencing data. All gene names used are according to IMGT nomenclature. Data visualization and TCR repertoire comparison were done in R (version 3.1.2). The IMGT/Junction Analysis tool was used to analyze in detail the CDR3 V-D-J and V-J junctions.
Human cell phenotypic analysis. Peripheral blood mononuclear cells (PBMCs) were isolated by density centrifugation and resuspended in RPMI 1640 supplemented with penicillin, streptomycin, and l-glutamine along with 10% fetal calf serum. PBMCs rested for 1 hour in the media at 37°C. Subsequently, cells were costimulated with PMA and ionomycin (Calbiochem) plus brefeldin A (Sigma-Aldrich). Cells were incubated for 5 hours at 37°C. Following incubation, PBMCs were washed in PBS containing 0.5% BSA and stained for extracellular markers including Live/Dead Fixable Blue Viability Dye (Invitrogen); CD3 (clone UCHT1), CD27 (clone L128) (both BD Biosciences); CD4 (clone RPA-T4, BioLegend); CD45RO (clone UCHL1, Beckman Coulter); TCRγδ (clone REA591), Vδ1 (clone REA173), and Vδ2 (clone 123R3) (all Miltenyi Biotec). PBMCs were then fixed and permeabilized using BD Biosciences Cytofix/Cytoperm Kit. Finally, PBMCs were stained for intracellular cytokine production of TNF (clone mAb 11 RUO) IFN-γ (clone B27) (both BD Biosciences), and IL-17A (clone eBio64CAP17, eBioscience) and analyzed by flow cytometry and using FlowJo software (Tree Star).
Statistics. Statistical analysis of the RNA-seq data is described above. For all other experiments, data for multiple comparisons were compared using 2-way ANOVA, and data from single comparisons were compared using 2-tailed Student’s t test. In certain instances, Bonferroni’s correction was applied to account for multiple comparisons, as indicated in the figure legends. All statistical analyses were calculated using Prism software (GraphPad). Data are presented as mean ± SEM, and values of P < 0.05 were considered to be statistically significant.
Study approval. All mice were bred and maintained under specific pathogen–free conditions at an animal facility accredited by the American Association for the Accreditation of Laboratory Animal Care (AAALAC) at Johns Hopkins and housed according to procedures described in the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011). All animal studies were approved by the Johns Hopkins University Animal Care and Use Committee. All human subject samples were gathered at the NIH Clinical Center via protocol NCT00001355, approved by the National Institute of Allergy and Infectious Diseases Institutional Review Board. These included healthy human subjects (n = 6) and individuals with genetically confirmed IRAK4 deficiency (n = 2) or CGD (n = 4 individuals). Both individuals with IRAK4 deficiency had a history of recurrent S. aureus skin infections, 3 of the 4 individuals with CGD had a history of recurrent S. aureus skin infections, and all 4 individuals with CGD had a history of an S. aureus liver abscess. All samples were collected from adults, with the exception of the first of 2 samples from an IRAK4-deficient patient, which were drawn when she was 17 (a sample that was collected toward the end of her predilection for recurrent infection). All patients gave informed consent.
-
Author contributions
CAD, BLP, AIM, AAM, ONF, HL, NKA, DBL, YW, RVO, SKL, MCM, SSC, AGA, LS May, LGM, MRY, SIS, JDM, EM, and LS Miller performed experiments and analyzed data. AFF, SMH, and JDM evaluated patients and obtained samples. CAD, AIM, AAM, ONF, LS May, LGM, MRY, SIS, JDM, EM, and LS Miller conceived the study, designed experiments, interpreted data and wrote the manuscript.
-
Acknowledgments
This work was supported by grants R01AR069502 (to L.S. Miller), R21AI126896 (to L.S. Miller), 1DP2OD008752 (to EM), U01AI124319 (to MRY), and R01AI129302 (to SIS) and in part by the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases of the US NIH (to SMH, AFF, and JDM) and an early career award from the Burroughs Wellcome Fund (to EM). The content is solely the responsibility of the authors and does not necessarily represent the official views of the US NIH. We thank Tammy Kielian (University of Nebraska) for providing the USA300 LAC::lux strain and Yoichiro Iwakura (University of Tokyo) for generously providing the IL-1β–/– mice.
Address correspondence to: Lloyd S. Miller, Johns Hopkins Department of Dermatology, Cancer Research Building II, Suite 205, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.955.8662; Email: lloydmiller@jhmi.edu.
-
Footnotes
Conflict of interest: L.S. Miller reports grant support from MedImmune, Regeneron Pharmaceuticals, Moderna Therapeutics, and Pfizer, which are developing therapeutics and vaccines against Staphylococcus aureus and other pathogens. M.R. Yeaman is a shareholder of NovaDigm Therapeutics, which is developing new vaccines, including NDV-3 targeting S. aureus and other pathogens.
Reference information: J Clin Invest. 2018;128(3):1026–1042.https://doi.org/10.1172/JCI96481.
-
References
- Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28(3):603–661.
- Montgomery CP, David MZ, Daum RS. Host factors that contribute to recurrent staphylococcal skin infection. Curr Opin Infect Dis. 2015;28(3):253–258.
- Holtfreter S, Kolata J, Bröker BM. Towards the immune proteome of Staphylococcus aureus - The anti-S. aureus antibody response. Int J Med Microbiol. 2010;300(2-3):176–192.
- Kolata JB, et al. The fall of a dogma? unexpected high t-cell memory response to Staphylococcus aureus in humans. J Infect Dis. 2015;212(5):830–838.
- Fowler VG, Proctor RA. Where does a Staphylococcus aureus vaccine stand? Clin Microbiol Infect. 2014;20 Suppl 5:66–75.View this article via: PubMed Google Scholar
- Miller LS, Cho JS. Immunity against Staphylococcus aureus cutaneous infections. Nat Rev Immunol. 2011;11(8):505–518.
- Bouma G, et al. Impaired neutrophil migration and phagocytosis in IRAK-4 deficiency. Br J Haematol. 2009;147(1):153–156.
- Picard C, et al. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science. 2003;299(5615):2076–2079.
- von Bernuth H, et al. Pyogenic bacterial infections in humans with MyD88 deficiency. Science. 2008;321(5889):691–696.
- Lévy R, et al. Genetic, immunological, and clinical features of patients with bacterial and fungal infections due to inherited IL-17RA deficiency. Proc Natl Acad Sci U S A. 2016;113(51):E8277–E8285.
- Milner JD, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452(7188):773–776.
- Puel A, et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science. 2011;332(6025):65–68.
- Cho JS, et al. Neutrophil-derived IL-1β is sufficient for abscess formation in immunity against Staphylococcus aureus in mice. PLoS Pathog. 2012;8(11):e1003047.
- Miller LS, et al. MyD88 mediates neutrophil recruitment initiated by IL-1R but not TLR2 activation in immunity against Staphylococcus aureus. Immunity. 2006;24(1):79–91.
- Chan LC, et al. Nonredundant roles of interleukin-17A (IL-17A) and IL-22 in murine host defense against cutaneous and hematogenous infection due to methicillin-resistant Staphylococcus aureus. Infect Immun. 2015;83(11):4427–4437.
- Cho JS, et al. IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J Clin Invest. 2010;120(5):1762–1773.
- Gaidamakova EK, et al. Preserving immunogenicity of lethally irradiated viral and bacterial vaccine epitopes using a radio- protective Mn2+-Peptide complex from Deinococcus. Cell Host Microbe. 2012;12(1):117–124.
- Montgomery CP, Daniels M, Zhao F, Alegre ML, Chong AS, Daum RS. Protective immunity against recurrent Staphylococcus aureus skin infection requires antibody and interleukin-17A. Infect Immun. 2014;82(5):2125–2134.
- Picard C, et al. Clinical features and outcome of patients with IRAK-4 and MyD88 deficiency. Medicine (Baltimore). 2010;89(6):403–425.
- Patel DD, Kuchroo VK. Th17 cell pathway in human immunity: lessons from genetics and therapeutic interventions. Immunity. 2015;43(6):1040–1051.
- Sampedro GR, et al. Targeting Staphylococcus aureus α-toxin as a novel approach to reduce severity of recurrent skin and soft-tissue infections. J Infect Dis. 2014;210(7):1012–1018.
- Tsuda Y, Takahashi H, Kobayashi M, Hanafusa T, Herndon DN, Suzuki F. Three different neutrophil subsets exhibited in mice with different susceptibilities to infection by methicillin-resistant Staphylococcus aureus. Immunity. 2004;21(2):215–226.
- Chan LC, et al. Innate immune memory contributes to host defense against recurrent skin and skin structure infections caused by methicillin-resistant Staphylococcus aureus. Infect Immun. 2017;85(2):e00876-16. View this article via: PubMed Google Scholar
- Ramírez-Valle F, Gray EE, Cyster JG. Inflammation induces dermal Vγ4+ γδT17 memory-like cells that travel to distant skin and accelerate secondary IL-17-driven responses. Proc Natl Acad Sci USA. 2015;112(26):8046–8051.
- Alegre ML, et al. Impact of Staphylococcus aureus USA300 colonization and skin infections on systemic immune responses in humans. J Immunol. 2016;197(4):1118–1126.
- Utay NS, et al. MRSA infections in HIV-infected people are associated with decreased MRSA-specific Th1 immunity. PLoS Pathog. 2016;12(4):e1005580.
- Zielinski CE, et al. Pathogen-induced human TH17 cells produce IFN-γ or IL-10 and are regulated by IL-1β. Nature. 2012;484(7395):514–518.
- Sheridan BS, et al. γδ T cells exhibit multifunctional and protective memory in intestinal tissues. Immunity. 2013;39(1):184–195.
- Cai Y, et al. Differential developmental requirement and peripheral regulation for dermal Vγ4 and Vγ6T17 cells in health and inflammation. Nat Commun. 2014;5:3986. View this article via: PubMed Google Scholar
- Pantelyushin S, et al. Rorγt+ innate lymphocytes and γδ T cells initiate psoriasiform plaque formation in mice. J Clin Invest. 2012;122(6):2252–2256.
- Sumaria N, et al. Cutaneous immunosurveillance by self-renewing dermal γδ T cells. J Exp Med. 2011;208(3):505–518.
- Murphy AG, O’Keeffe KM, Lalor SJ, Maher BM, Mills KH, McLoughlin RM. Staphylococcus aureus infection of mice expands a population of memory γδ T cells that are protective against subsequent infection. J Immunol. 2014;192(8):3697–3708.
- Bonneville M, O’Brien RL, Born WK. γδ T cell effector functions: a blend of innate programming and acquired plasticity. Nat Rev Immunol. 2010;10(7):467–478.
- Lalor SJ, McLoughlin RM. Memory γδ T cells-newly appreciated protagonists in infection and immunity. Trends Immunol. 2016;37(10):690–702.
- Vantourout P, Hayday A. Six-of-the-best: unique contributions of γδ T cells to immunology. Nat Rev Immunol. 2013;13(2):88–100.
- Brady RA, Bruno VM, Burns DL. RNA-seq analysis of the host response to Staphylococcus aureus skin and soft tissue infection in a mouse model. PLoS One. 2015;10(4):e0124877.
- Holland SM. Chronic granulomatous disease. Clin Rev Allergy Immunol. 2010;38(1):3–10.
- Chandran SS, Verhoeven D, Teijaro JR, Fenton MJ, Farber DL. TLR2 engagement on dendritic cells promotes high frequency effector and memory CD4 T cell responses. J Immunol. 2009;183(12):7832–7841.
- Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4(12):1191–1198.
- Nielsen MM, Witherden DA, Havran WL. γδ T cells in homeostasis and host defence of epithelial barrier tissues. Nat Rev Immunol. 2017;17(12):733–745.
- Martin B, Hirota K, Cua DJ, Stockinger B, Veldhoen M. Interleukin-17-producing γδ T cells selectively expand in response to pathogen products and environmental signals. Immunity. 2009;31(2):321–330.
- Barin JG, et al. Collaborative interferon-γ and interleukin-17 signaling protects the oral mucosa from Staphylococcus aureus. Am J Pathol. 2016;186(9):2337–2352.
- Asarnow DM, Kuziel WA, Bonyhadi M, Tigelaar RE, Tucker PW, Allison JP. Limited diversity of gamma delta antigen receptor genes of Thy-1+ dendritic epidermal cells. Cell. 1988;55(5):837–847.
- Itohara S, et al. Homing of a gamma delta thymocyte subset with homogeneous T-cell receptors to mucosal epithelia. Nature. 1990;343(6260):754–757.
- Muñoz-Ruiz M, Sumaria N, Pennington DJ, Silva-Santos B. Thymic determinants of γδ T cell differentiation. Trends Immunol. 2017;38(5):336–344.
- Havran WL, Chien YH, Allison JP. Recognition of self antigens by skin-derived T cells with invariant γδ antigen receptors. Science. 1991;252(5011):1430–1432.
- Komori HK, et al. Cutting edge: dendritic epidermal γδ T cell ligands are rapidly and locally expressed by keratinocytes following cutaneous wounding. J Immunol. 2012;188(7):2972–2976.
- Heyborne KD, Cranfill RL, Carding SR, Born WK, O’Brien RL. Characterization of gamma delta T lymphocytes at the maternal-fetal interface. J Immunol. 1992;149(9):2872–2878.View this article via: PubMed Google Scholar
- Reardon C, Lefrancois L, Farr A, Kubo R, O’Brien R, Born W. Expression of gamma/delta T cell receptors on lymphocytes from the lactating mammary gland. J Exp Med. 1990;172(4):1263–1266.
- Paget C, et al. CD3bright signals on γδ T cells identify IL-17A-producing Vγ6Vδ1+ T cells. Immunol Cell Biol. 2015;93(2):198–212.
- Conti HR, et al. Oral-resident natural Th17 cells and γδ T cells control opportunistic Candida albicans infections. J Exp Med. 2014;211(10):2075–2084.
- Misiak A, Wilk MM, Raverdeau M, Mills KH. IL-17-producing innate and pathogen-specific tissue resident memory γδ T cells expand in the lungs of Bordetella pertussis-infected mice. J Immunol. 2017;198(1):363–374.
- Okamoto Yoshida Y, et al. Essential role of IL-17A in the formation of a mycobacterial infection-induced granuloma in the lung. J Immunol. 2010;184(8):4414–4422.
- Davey MS, et al. Clonal selection in the human Vδ1 T cell repertoire indicates γδ TCR-dependent adaptive immune surveillance. Nat Commun. 2017;8:14760. View this article via: PubMed Google Scholar
- Dimova T, et al. Effector Vγ9Vδ2 T cells dominate the human fetal γδ T-cell repertoire. Proc Natl Acad Sci USA. 2015;112(6):E556–E565.
- Papotto PH, Ribot JC, Silva-Santos B. IL-17+ γδ T cells as kick-starters of inflammation. Nat Immunol. 2017;18(6):604–611.
- Karauzum H, Haudenschild CC, Moore IN, Mahmoudieh M, Barber DL, Datta SK. Lethal CD4 T cell responses induced by vaccination against Staphylococcus aureus bacteremia. J Infect Dis. 2017;215(8):1231–1239.
- Sanchez M, et al. O-acetylation of peptidoglycan limits helper T cell priming and permits Staphylococcus aureus reinfection. Cell Host Microbe. 2017;22(4):543–551.e4.
- Boyden LM, et al. Skint1, the prototype of a newly identified immunoglobulin superfamily gene cluster, positively selects epidermal gammadelta T cells. Nat Genet. 2008;40(5):656–662.
- Keyes BE, et al. Impaired epidermal to dendritic T cell signaling slows wound repair in aged skin. Cell. 2016;167(5):1323–1338.e14.
- Marusina AI, et al. CD4+ virtual memory: Antigen-inexperienced T cells reside in the naive, regulatory, and memory T cell compartments at similar frequencies, implications for autoimmunity. J Autoimmun. 2017;77:76–88.
-
Version history
- Version 1 (February 5, 2018): Electronic publication
- Version 2 (March 1, 2018): Print issue publication



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)














