Citation Information: J Clin Invest. 2016;126(4):1592-1602. https://doi.org/10.1172/JCI85908.
Abstract
The alternatively spliced products of
Authors
John M. Lee, Chika Nobumori, Yiping Tu, Catherine Choi, Shao H. Yang, Hea-Jin Jung, Timothy A. Vickers, Frank Rigo, C. Frank Bennett, Stephen G. Young, Loren G. Fong
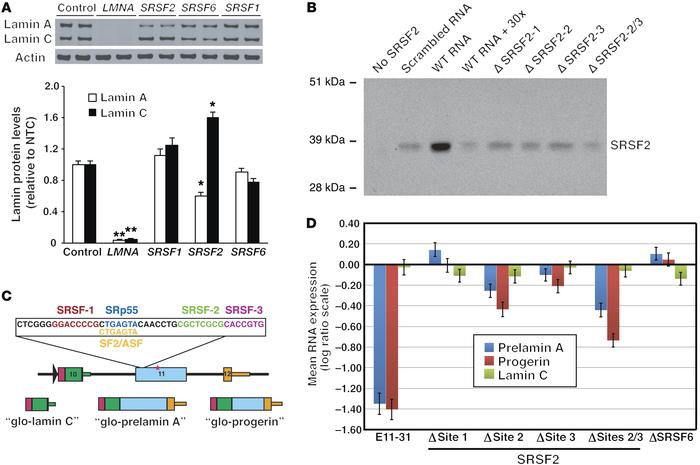
Figure 4
SRSF2 modulates



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)