Advertisement
Research ArticleVascular biology Free access | 10.1172/JCI84652
CHD4-regulated plasmin activation impacts lymphovenous hemostasis and hepatic vascular integrity
Patrick L. Crosswhite,1 Joanna J. Podsiadlowska,1 Carol D. Curtis,1 Siqi Gao,1 Lijun Xia,1,2 R. Sathish Srinivasan,1,3 and Courtney T. Griffin1,3
1Cardiovascular Biology Research Program, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma, USA.
2Department of Biochemistry and Molecular Biology and
3Department of Cell Biology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma, USA.
Address correspondence to: Courtney T. Griffin, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma 73104, USA. Phone: 405.271.7073; E-mail: courtney-griffin@omrf.org.
Find articles by Crosswhite, P. in: JCI | PubMed | Google Scholar
1Cardiovascular Biology Research Program, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma, USA.
2Department of Biochemistry and Molecular Biology and
3Department of Cell Biology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma, USA.
Address correspondence to: Courtney T. Griffin, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma 73104, USA. Phone: 405.271.7073; E-mail: courtney-griffin@omrf.org.
Find articles by Podsiadlowska, J. in: JCI | PubMed | Google Scholar
1Cardiovascular Biology Research Program, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma, USA.
2Department of Biochemistry and Molecular Biology and
3Department of Cell Biology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma, USA.
Address correspondence to: Courtney T. Griffin, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma 73104, USA. Phone: 405.271.7073; E-mail: courtney-griffin@omrf.org.
Find articles by Curtis, C. in: JCI | PubMed | Google Scholar
1Cardiovascular Biology Research Program, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma, USA.
2Department of Biochemistry and Molecular Biology and
3Department of Cell Biology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma, USA.
Address correspondence to: Courtney T. Griffin, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma 73104, USA. Phone: 405.271.7073; E-mail: courtney-griffin@omrf.org.
Find articles by Gao, S. in: JCI | PubMed | Google Scholar
1Cardiovascular Biology Research Program, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma, USA.
2Department of Biochemistry and Molecular Biology and
3Department of Cell Biology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma, USA.
Address correspondence to: Courtney T. Griffin, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma 73104, USA. Phone: 405.271.7073; E-mail: courtney-griffin@omrf.org.
Find articles by
Xia, L.
in:
JCI
|
PubMed
|
Google Scholar
|

1Cardiovascular Biology Research Program, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma, USA.
2Department of Biochemistry and Molecular Biology and
3Department of Cell Biology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma, USA.
Address correspondence to: Courtney T. Griffin, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma 73104, USA. Phone: 405.271.7073; E-mail: courtney-griffin@omrf.org.
Find articles by Srinivasan, R. in: JCI | PubMed | Google Scholar
1Cardiovascular Biology Research Program, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma, USA.
2Department of Biochemistry and Molecular Biology and
3Department of Cell Biology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma, USA.
Address correspondence to: Courtney T. Griffin, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma 73104, USA. Phone: 405.271.7073; E-mail: courtney-griffin@omrf.org.
Find articles by Griffin, C. in: JCI | PubMed | Google Scholar
Published May 3, 2016 - More info
J Clin Invest. 2016;126(6):2254–2266. https://doi.org/10.1172/JCI84652.
Copyright © 2016, American Society for Clinical Investigation
Received: September 14, 2015; Accepted: March 10, 2016
-
Abstract
The chromatin-remodeling enzyme CHD4 maintains vascular integrity at mid-gestation; however, it is unknown whether this enzyme contributes to later blood vessel or lymphatic vessel development. Here, we addressed this issue in mice harboring a deletion of Chd4 specifically in cells that express lymphatic vessel endothelial hyaluronan receptor 1 (LYVE1), which include lymphatic endothelial cells (LECs) and liver sinusoidal endothelial cells. Chd4 mutant embryos died before birth and exhibited severe edema, blood-filled lymphatics, and liver hemorrhage. CHD4-deficient embryos developed normal lymphovenous (LV) valves, which regulate the return of lymph to the blood circulation; however, these valves lacked the fibrin-rich thrombi that prevent blood from entering the lymphatic system. Transcripts of the urokinase plasminogen activator receptor (uPAR), which facilitates activation of the fibrin-degrading protease plasmin, were upregulated in Chd4 mutant LYVE1+ cells, and plasmin activity was elevated near the LV valves. Genetic reduction of the uPAR ligand urokinase prevented degradation of fibrin-rich thrombi at the LV valves and largely resolved the blood-filled lymphatics in Chd4 mutants. Urokinase reduction also ameliorated liver hemorrhage and prolonged embryonic survival by reducing plasmin-mediated extracellular matrix degradation around sinusoidal blood vessels. These results highlight the susceptibility of LV thrombi and liver sinusoidal vessels to plasmin-mediated damage and demonstrate the importance of CHD4 in regulating embryonic plasmin activation after mid-gestation.
-
Introduction
The circulatory system in mammals is divided into two distinct vascular networks: the blood and lymphatic systems. The blood system transports blood through a high-pressure, closed system that delivers oxygen and nutrients to the tissues. Hydrostatic pressure generated by the heart pushes water from blood out of tissue capillaries, and water that does not reenter capillaries through osmosis accumulates as interstitial fluid (1). A key role of the lymphatic system is to collect excessive interstitial fluid from the tissues and return it to the blood system. Lymphovenous (LV) valves are important gatekeepers at the junction between the blood and lymphatic systems that help facilitate this return of interstitial fluid collected by the lymphatics back to the blood circulation. These valves form at the sites where the thoracic and right lymphatic ducts intersect with the subclavian and internal jugular veins. In their absence, interstitial fluid accumulates in tissues, and severe edema ensues (2–4).
Despite the importance of LV valves in mediating systemic fluid balance, these valves are not sufficient to prevent blood from backflowing into the lymphatic system. Platelet thrombi at the LV valves are also required for maintaining the separation of the blood and lymphatic systems (5). Platelet aggregation in the lymphatic system is initiated when the platelet receptor C-type lectin-like receptor 2 (CLEC2) interacts with the mucin-type transmembrane protein podoplanin (PDPN) on the surface of lymphatic endothelial cells (LECs) (6). Murine embryos with genetic deletion of Clec2 fail to form platelet aggregates at the LV valves and subsequently undergo retrograde flow of blood into the thoracic duct and downstream lymphatic vessels (5). Importantly, LV thrombi likely continue to play important roles in blood/lymph separation after birth, since neonatal mice injected with anti-CLEC2 antibodies and mature mice reconstituted with Clec2–/– hematopoietic cells develop blood-filled lymphatics soon after treatment (5). Questions remain, however, about the regulation of LV thrombi, since large and stable clots would presumably impede lymph flow through the LV valves. Therefore, LV thrombus formation and degradation must be tightly coordinated in order to facilitate lymph flow into the blood system while preventing retrograde flow of blood into the lymphatic system. Because LV thrombi stain strongly for fibrin, which is generated on activated platelet surfaces by thrombin and acts to stabilize platelet clots (7), fibrinolysis has been proposed as a mechanism for regulating LV thrombus degradation (5).
The serine protease plasmin is a key mediator of fibrinolysis and thrombus dissolution (8). Plasmin activation is triggered by the tissue-type and urokinase-type plasminogen activators (tPA and uPA). Mid-gestational embryonic uPA and uPA receptor (uPAR) expression are regulated by the chromodomain helicase DNA-binding 4 (CHD4, also known as Mi-2β) protein, an ATPase within the nucleosome remodeling deacetylase (NuRD) chromatin-remodeling complex (9). Chromatin-remodeling complexes such as NuRD impact target gene expression by transiently disrupting DNA-histone contacts and thereby altering the accessibility of promoter regions to transcriptional regulators (10, 11). Embryos depleted for Chd4 in vascular endothelial cells die from vascular rupture at mid-gestation due to excessive uPA and uPAR transcription that leads to plasmin activation and degradation of extracellular matrix components, which — like fibrin — are cleaved by plasmin (9). However, the lethality of these embryos prevented assessment of whether CHD4 regulates plasmin activation and other critical processes later in vascular development.
We now report that embryos lacking CHD4 in lymphatic vessel endothelial hyaluronan receptor 1–positive (LYVE1+) cells die during embryonic development, with severe edema, blood-filled lymphatics, and liver hemorrhage. We show that loss of CHD4 in LYVE1+ LECs increases plasmin activation in the lymphatic system, which degrades fibrin-rich thrombi at the LV valves and allows blood to enter the lymphatic system. Simultaneously, loss of CHD4 in LYVE1+ hepatic sinusoidal endothelial cells causes excessive plasmin activity, degradation of the vascular extracellular matrix component laminin, hepatic vascular rupture, and embryonic lethality. Our data provide insights into the importance of CHD4-regulated plasmin activation after mid-gestation and the detrimental impact of excessive plasmin activity on lymphatic and hepatic vascular development.
-
Results
Chd4fl/fl VE-cadherin-Cre+ embryos die by E16.5, with blood-filled lymphatic vessels and edema. Because Chd4fl/fl Tie2-Cre+ embryos die by E11.5 from vascular rupture and hemorrhage (9), we sought alternative genetic approaches for assessing the role of CHD4 in later vascular development. We first crossed Chd4fl/fl mice onto the VE-cadherin-Cre transgenic background, in which Cre recombinase is expressed in developing endothelial cells but is not fully penetrant until E14.5 (12). We found no surviving Chd4fl/fl VE-cadherin-Cre+ offspring at weaning from crosses that were designed to generate 25% of this genotype (χ2 = 26.2, P < 0.001, Table 1). When we assessed embryos from timed matings, we found evidence of edema and superficial blood-filled vessels in Chd4fl/fl VE-cadherin-Cre+ mutants by E14.5 (Figure 1, A and D). These phenotypes became more pronounced in Chd4fl/fl VE-cadherin-Cre+ mutants through E16.5, when lethality occurred (Figure 1, B, C, E, and F). Because the edema and superficial blood-filled vessels we saw in Chd4fl/fl VE-cadherin-Cre+ mutants were reminiscent of phenotypes seen in embryos with defective lymphatic vascular development (13), and because VE-cadherin-Cre is expressed in LECs (Supplemental Figure 1A and ref. 12), we questioned whether Chd4fl/fl VE-cadherin-Cre+ embryos harbored abnormal lymphatic vasculature. Immunostaining analysis of the jugular lymph sacs and dermal lymphatic vessels revealed dilated and blood-filled lymphatic vasculature in Chd4fl/fl VE-cadherin-Cre+ embryos by E14.5 (Figure 1, G–J). These phenotypes drove us to seek a more specific genetic assay for determining the role of CHD4 in developing lymphatic vessels.
 Figure 1
Figure 1Chd4fl/fl VE-cadherin-Cre+ and Chd4fl/fl Lyve1-Cre+ embryos display edema and blood-filled lymphatics before death. (A-F) Littermate control (A–C) and Chd4fl/fl VE-cadherin-Cre+ (D–F) embryos were photographed from E14.5 to E16.5. n = 10 embryos per genotype per time point. Mutants displayed edema (white arrows) and prominent superficial blood-filled vessels (white arrowheads) as early as E14.5 and died by E16.5. (G–J) Sections from E14.5 control (G and H) and Chd4fl/fl VE-cadherin-Cre+ (I and J) embryos were immunostained for the red blood cell marker Ter119 (red) and the lymphatic marker LYVE1 (green), and nuclei were counterstained with Hoechst (blue). The Chd4fl/fl VE-cadherin-Cre+ embryos had enlarged, blood-filled jugular lymph sacs (JLS; I, white arrow) and dermal lymphatic vessels (DLV; J, white arrow). n = 3 embryos per genotype. (K–P) Littermate control (K–M) and Chd4fl/fl Lyve1-Cre+ (N–P) embryos were photographed from E13.5 to E14.5. Mutants displayed edema (white arrows) as early as E13.5 and superficial blood-filled vessels (white arrowheads) by early E14.5, and died by late E14.5. n = 10 embryos per genotype per time point. (Q and R) Sections from E14.5 control (Q) and Chd4fl/fl Lyve1-Cre+ (R) embryos were immunostained for LYVE1 (red), CHD4 (green), and Hoechst (blue) to assess CHD4 expression in LECs (white arrows). Insets show 5× magnified images of LECs in boxed regions. n = 3 embryos per genotype. (S and T) Sections from late E14.5 control (S) and Chd4fl/fl Lyve1-Cre+ (T) embryos were stained with H&E. Red blood cells (black arrows) were detected in the enlarged jugular lymph sac (JLS) of mutant embryos. n = 3 embryos per genotype. For A–F and K–P, embryos at each time point were imaged at the same magnification. Scale bars: 100 μm (G–J, S, and T); 50 μm (Q and R). JV, jugular vein; CA, carotid artery.
Chd4fl/fl Lyve1-Cre+ embryos die by E14.5, with blood-filled lymphatic vessels and edema. We next deleted Chd4 from LECs using the Lyve1-Cre+ knock-in line (14). Chd4fl/fl females were mated with Chd4fl/+ Lyve1-Cre+ males, and progeny from 13 litters were genotyped and scored at weaning. No live Chd4fl/fl Lyve1-Cre+ mice were detected (χ2 = 27.5, P < 0.001, Table 2). When we performed timed matings to determine the time at which Chd4fl/fl Lyve1-Cre+ embryos died, we found live but slightly pale and edematous mutant embryos at E13.5 (Figure 1, K and N). One day later, by late E14.5, we found dead Chd4fl/fl Lyve1-Cre+ embryos with pronounced edema and superficial blood-filled vessels (Figure 1, L, M, O, and P). We confirmed that lymphatic vessels form and express LYVE1 in Chd4fl/fl Lyve1-Cre+ embryos (Supplemental Figure 2) and that CHD4 is efficiently deleted from Chd4fl/fl Lyve1-Cre+ LECs by E14.5 (Figure 1, Q and R). Histological analysis of Chd4fl/fl Lyve1-Cre+ embryos revealed minor amounts of blood in jugular lymph sacs of E12.5 and E13.5 embryos (data not shown) and larger quantities of blood in dilated jugular lymph sacs and dermal lymphatics prior to lethality at E14.5 (Supplemental Figure 2 and Figure 1, S and T). Thus, Chd4fl/fl Lyve1-Cre+ embryos died with gross and histological phenotypic similarities to Chd4fl/fl VE-cadherin-Cre+ embryos, but the cause of embryonic lethality in both mutant lines was not obvious, since edema and blood-filled lymphatics do not always correlate with embryonic death (15–20).
Chd4fl/fl Lyve1-Cre+ embryos have structurally normal LV valves. LV valves help prevent blood from aberrantly entering the lymphatic system, and their development overlaps in timing with the manifestation of our Chd4fl/fl Lyve1-Cre+ phenotype (2, 4). Therefore, we examined the mutants to assess their LV valve morphology. First, we confirmed that Lyve1-Cre is active around the LV valves at E14.5 using a Cre reporter line (Supplemental Figure 1B). Next, using coronal sections and H&E staining, we determined that Chd4fl/fl Lyve1-Cre+ embryos developed LV valves similar to those seen in their control littermates (Figure 2, A–D). Moreover, E13.5 Chd4fl/fl Lyve1-Cre+ LV valves expressed the transcription factor PROX1, which is critical for LV valve formation (2, 4, 21), at a level comparable to that seen in littermate controls (Figure 2, E and F). Therefore, LV valve development is not impaired by Chd4 deletion in LECs, so an LV valve deficit is not the cause of blood-filled lymphatics in Chd4fl/fl Lyve1-Cre+ embryos.
 Figure 2
Figure 2Chd4fl/fl Lyve1-Cre+ embryos have structurally normal LV valves. (A–D) Coronal sections of early (E14.5) littermate control (A and C) and Chd4fl/fl Lyve1-Cre+ (B and D) embryos were stained with H&E. Control and Chd4fl/fl Lyve1-Cre+ embryos displayed structurally similar LV valves (circles). n = 6 embryos per genotype. (E and F) Sections of E13.5 control (E) and Chd4fl/fl Lyve1-Cre+ (F) LV valves (white arrows) were immunostained for PROX1 (red) and LYVE1 (green) and counterstained with Hoechst (blue). Control and mutant LV valves expressed the valve marker PROX1 to a similar degree. n = 3 embryos per genotype. Scale bars: 100 μm (A and B); 50 μm (C–F).
Chd4fl/fl Lyve1-Cre+ embryos lack fibrin-rich LV thrombi. Since fibrin-rich thrombi at the LV valves help prevent blood from entering the lymphatic circulation (5), we next analyzed LV thrombi in control and Chd4fl/fl Lyve1-Cre+ mutants by immunostaining for fibrin. While control embryos showed robust fibrin staining at the LV valves (Figure 3, A and C, n = 4), we were unable to detect fibrin at the LV valves of Chd4fl/fl Lyve1-Cre+ embryos (Figure 3, B and D, n = 7). We suspected that the lack of fibrin-rich LV thrombi in Chd4fl/fl Lyve1-Cre+ embryos could explain the aberrant blood we observed in their lymphatic vessels.
 Figure 3
Figure 3Chd4fl/fl Lyve1-Cre+ embryos lack fibrin-rich thrombi at their LV valves and have excessive lymphatic plasmin activation. (A–D) Sections of LV valves from E14.5 control (A and C) and Chd4fl/fl Lyve1-Cre+ (B and D) embryos were immunostained for fibrin (red) and LYVE1 (green) and counterstained with Hoechst (blue). Fibrin-rich thrombi (white arrows) were detected at the control LV valves, but no comparable thrombi were detected at the mutant LV valves (white arrowheads). n = 5 embryos per genotype. (E) LYVE1+ cells were purified from E13.5 littermate control and Chd4fl/fl Lyve1-Cre+ embryos. RNA was isolated, and qPCR was used to determine the expression of selected genes. n = 4 embryos per genotype. Values expressed as relative normalized expression are mean ± SEM. *P = 0.0120, calculated by Student’s t test. (F and G) Unfixed sections of jugular lymph sacs (outlined in white) from E12.5 control (F) and Chd4fl/fl Lyve1-Cre+ (G) embryos were subjected to in situ zymography for detection of plasmin activity. Sections were overlaid with quenched fluorescent casein, a plasmin substrate that fluoresces upon cleavage. In the presence of exogenous plasminogen, plasmin activity (green fluorescence) was substantially higher around the LECs (white arrows) lining the Chd4fl/fl Lyve1-Cre+ lymph sacs versus the control lymph sacs. n = 3 embryos per genotype. Scale bars: 50 μm (A–D); 100 μm (F and G).
We recognized that the deficit of LV thrombi in our Chd4fl/fl Lyve1-Cre+ embryos could result from failed thrombus formation or from excessive thrombus degradation. Since LV thrombi were difficult for us to detect prior to E14.5, we resorted to indirect methods for determining whether thrombi could form in Chd4fl/fl Lyve1-Cre+ embryos. LV thrombus formation is initiated when platelets expressing CLEC2 interact with LECs expressing PDPN (5). Lyve1-Cre expression in embryonic megakaryocytes or platelets has not been reported, so we did not expect excision of Chd4 in platelets to cause the LV thrombus deficit in Chd4fl/fl Lyve1-Cre+ embryos. Nevertheless, we addressed a requirement for platelet CHD4 in LV thrombus formation by crossing Chd4fl/fl mice onto the megakaryocyte/platelet-specific PF4-Cre transgenic line (22). Expected numbers of Chd4fl/fl PF4-Cre+ mice were born and survived through weaning (Supplemental Table 1), and Chd4fl/fl PF4-Cre+ embryos displayed no evidence of edema or blood-filled lymphatics at E14.5 (Supplemental Figure 3, A and B). These results indicated that CHD4 expression in platelets is not required for embryonic LV thrombus formation. We also addressed the possibility that the deficit of LV thrombi in Chd4fl/fl Lyve1-Cre+ embryos resulted from a downregulation of PDPN on LECs. We isolated LYVE1+ cells from E13.5 control and Chd4fl/fl Lyve1-Cre+ embryos but saw no significant difference in Pdpn expression by quantitative PCR (qPCR) (Figure 3E). Together, these data suggested that Chd4fl/fl Lyve1-Cre+ embryos express the platelet and LEC components required to initiate LV thrombus formation. Therefore, we considered mechanisms by which CHD4 might regulate LV thrombus dissolution.
The serine protease plasmin degrades fibrin-rich thrombi (23). We questioned whether elevated plasmin in Chd4fl/fl Lyve1-Cre+ embryos could be responsible for the lysis and disappearance of LV valve thrombi. To address this question, we first assessed whether plasmin activators or inhibitors were transcriptionally dysregulated in LYVE1+ cells isolated from Chd4fl/fl Lyve1-Cre+ embryos. We did not see significant changes in expression of the plasmin activators uPA and tPA (Figure 3E). Likewise, we saw no significant decrease in the plasmin inhibitor thrombospondin-1 (Thbs1) (Figure 3E). However, we found that uPAR was significantly upregulated in LYVE1+ cells isolated from Chd4fl/fl Lyve1-Cre+ embryos versus littermate controls (Figure 3E, P = 0.0120). This result is consistent with our previous finding that uPAR transcription is directly inhibited by the CHD4-NuRD complex in blood vessel endothelial cells at E10.5 (9).
uPAR is important for plasmin activation because it binds and localizes uPA to cell surfaces, where it facilitates cleavage of the plasmin zymogen plasminogen (24, 25). Therefore, upregulated uPAR expression could lead to elevated plasmin activation. To assess plasmin activity in the lymphatic vasculature of our control and mutant embryos, we performed in situ zymography on tissue sections of jugular lymph sacs from E12.5 embryos. Plasmin activity was detected with the quenched fluorescent plasmin substrate casein, which fluoresces upon cleavage. After addition of plasminogen, which normally circulates in plasma but must be added exogenously to tissue sections, plasmin activity was substantially elevated around LECs lining the jugular lymph sac of Chd4fl/fl Lyve1-Cre+ embryos but not around LECs of control embryos (Figure 3, F and G). These data supported our hypothesis that increased plasmin activity could cause the degradation of fibrin-rich thrombi at Chd4 mutant LV valves.
Genetic reduction of the plasmin activator uPA in Chd4fl/fl Lyve1-Cre+ embryos prevents degradation of the fibrin-rich thrombi at the LV valves, prevents blood-filled lymphatics, and prolongs survival. Since uPA is a potent activator of the fibrinolytic protease plasmin (24, 25), and since the uPA receptor, uPAR, is upregulated in LYVE1+ cells from Chd4fl/fl Lyve1-Cre+ embryos, we predicted that genetic reduction of uPA would decrease plasmin activity, prevent dissolution of the LV thrombi, and eliminate blood-filled lymphatics in Chd4fl/fl Lyve1-Cre+ embryos. We crossed Chd4fl/fl Lyve1-Cre+ embryos onto a heterozygous (uPA+/–) or knockout (uPA–/–) background. We were able to detect fibrin-rich thrombi at the LV valves of both uPA+/– Chd4fl/fl Lyve1-Cre+ and uPA–/– Chd4fl/fl Lyve1-Cre+ embryos (Figure 4, A–D, n = 2 for each genotype), which we had not been able to locate in Chd4fl/fl Lyve1-Cre+ embryos (Figure 3, B and D). Furthermore, blood-filled lymphatics were reduced on a uPA-deficient background. Histological examination of control embryos showed virtually no blood in the lymphatic circulation compared with 77% (17 of 22) of Chd4fl/fl Lyve1-Cre+ embryos (Table 3). However, only 33% (3 of 9) of uPA+/– Chd4fl/fl Lyve1-Cre+ embryos had blood in their lymphatics, and we detected only 1 (of 4) uPA–/– Chd4fl/fl Lyve1-Cre+ embryo with blood-filled lymphatics. Collectively, these data suggest that uPA reduction prevents LV valve clot degradation due to excessive plasmin activity and safeguards the lymphatic circulation from aberrant entry of blood.
 Figure 4
Figure 4Genetic reduction of the plasmin activator uPA in Chd4fl/fl Lyve1-Cre+ embryos prevents degradation of LV thrombi, prevents blood-filled lymphatics, and prolongs survival. (A–D) Sections of LV valves from E14.5 uPA+/– Chd4fl/fl Lyve1-Cre+ (A) and uPA–/– Chd4fl/fl Lyve1-Cre+ (B) embryos were immunostained for fibrin (red) and LYVE1 (green) and counterstained with Hoechst (blue). Fibrin-rich thrombi (white arrows) were detected at the LV valves in uPA-reduced mutants unlike in Chd4fl/fl Lyve1-Cre+ embryos without uPA reduction. Single-channel fibrin staining is shown in C and D. n = 2 embryos for A and C; n = 3 embryos for B and D. (E–H) Control and Chd4 mutant embryos with and without uPA reduction were dissected and photographed at E15.5, 1 day after Chd4fl/fl Lyve1-Cre+ embryos typically die. Of the Chd4fl/fl Lyve1-Cre+ embryos analyzed, 5 of 5 showed advanced resorption at E15.5 (F). However, death was delayed in 10 of 17 uPA+/– Chd4fl/fl Lyve1-Cre+ embryos (G) and in 6 of 8 uPA–/– Chd4fl/fl Lyve1-Cre+ embryos (H). All embryos were photographed at the same magnification. Scale bars: 50 μm (A and B).
We next dissected embryos at E15.5 to determine whether uPA reduction could rescue death in the Chd4fl/fl Lyve1-Cre+ mutants, which is typically observed at E14.5. Compared with control embryos (Figure 4E), all 5 Chd4fl/fl Lyve1-Cre+ embryos we assessed at E15.5 were severely necrotic and in advanced stages of resorption (Figure 4F). By contrast, 10 of 17 mutant embryos depleted of one uPA allele (uPA+/– Chd4fl/fl Lyve1-Cre+) showed delayed death at E15.5 (Figure 4G). Death was also delayed in 6 of 8 embryos depleted of both uPA alleles (uPA–/– Chd4fl/fl Lyve1-Cre+, Figure 4H). Although we detected a heartbeat in one uPA+/– Chd4fl/fl Lyve1-Cre+ embryo at E16.5, uPA reduction did not prevent the eventual death of Chd4fl/fl Lyve1-Cre+ embryos, since neither uPA+/– Chd4fl/fl Lyve1-Cre+ nor uPA–/– Chd4fl/fl Lyve1-Cre+ mice were recovered after birth or at weaning (Supplemental Table 2).
Fatal liver hemorrhage in Chd4fl/fl Lyve1-Cre+ mutants is ameliorated with uPA reduction. Since several lines of mutants with edema or blood-filled lymphatics survive embryonic development (15–20), we suspected that the lymphatic defects in our Chd4fl/fl Lyve1-Cre+ embryos were not the primary cause of lethality. Histological analysis of E14.5 control and mutant embryos revealed normal development of the heart, kidney, stomach, and intestine (Supplemental Figure 4). However, the livers from Chd4fl/fl Lyve1-Cre+ embryos showed substantial tissue deterioration and blood vessel hemorrhage — particularly at the periphery of the tissue — compared with livers from control embryos (Figure 5, A–F). Similar liver pathology was observed in E14.5 Chd4fl/fl VE-cadherin-Cre+ embryos prior to lethality at E16.5 (Supplemental Figure 5). The Lyve1-Cre recombinase we employed in this study was originally developed to excise target genes from lymphatic endothelium (14), but LYVE1 is also expressed on some blood vessel endothelial cells, including hepatic sinusoidal endothelial cells (14, 26–28). We confirmed Lyve1-Cre recombinase activity in E14.5 hepatic vessels with a Cre reporter (Supplemental Figure 6B). We also confirmed with immunostaining that CHD4 is expressed in cells lining LYVE1+ hepatic vessels and is reduced in LYVE1+ vessels within Chd4fl/fl Lyve1-Cre+ livers (Supplemental Figure 6, C–H). These LYVE1+/CHD4+ hepatic vessels are not likely to be lymphatics, because they do not express the LEC marker PDPN, which we found expressed robustly in LECs outside the liver but expressed only in mesothelial cells lining the liver lobes at E14.5 (Supplemental Figure 7). Therefore, we suspected that rupture of liver sinusoids and subsequent hemorrhage was the primary cause of death in Chd4fl/fl Lyve1-Cre+ embryos.
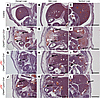 Figure 5
Figure 5Fatal liver hemorrhage in Chd4fl/fl Lyve1-Cre+ mutants is ameliorated with uPA reduction. Coronal sections (cut from dorsal to ventral) of paraffin-embedded E14.5 embryos were stained with H&E. Livers from control embryos (A–C) had compact parenchyma and normal vasculature, while livers from Chd4fl/fl Lyve1-Cre+ embryos (D–F) had substantial parenchymal deterioration (black arrows) and hemorrhage (white arrows). uPA+/– Chd4fl/fl Lyve1-Cre+ livers (G–I) showed less deterioration compared with Chd4fl/fl Lyve1-Cre+ livers (D–F), but parenchymal degradation and hemorrhage were still detected, particularly on the liver periphery. The benefit of uPA reduction was more obvious in livers from uPA–/– Chd4fl/fl Lyve1-Cre+ embryos (J–L), which had even less parenchymal degradation and hemorrhage throughout the entire organ and were closer in size to control livers. n = 3 embryos per genotype. Scale bars: 500 μm. L, liver; K, kidney; S, stomach.
We previously reported that deletion of Chd4 from blood vessel endothelial cells at mid-gestation results in vascular rupture due to excessive plasmin activation and degradation of the extracellular matrix surrounding the dorsal aortae and cardinal veins (9). Importantly, genetic reduction of the plasmin activator uPA partially rescues matrix degradation and vascular rupture in Chd4fl/fl Tie2-Cre+ embryos (9). We likewise observed reduced hemorrhage in E14.5 uPA+/– Chd4fl/fl Lyve1-Cre+ livers compared with Chd4fl/fl Lyve1-Cre+ livers (Figure 5, D–I). The benefit of genetic uPA reduction was even more evident in embryos lacking both alleles of uPA, since livers from uPA–/– Chd4fl/fl Lyve1-Cre+ embryos were even less hemorrhagic and showed no obvious developmental delay (Figure 5, J–L). Nevertheless, some hemorrhage was still evident in uPA–/– Chd4fl/fl Lyve1-Cre+ livers, which is consistent with the eventual death of these embryos.
Plasmin activity corresponds with vascular extracellular matrix degradation and hemorrhage in Chd4fl/fl Lyve1-Cre+ livers prior to embryonic lethality. Our genetic data indicated that Chd4 excision in LYVE1+ liver sinusoidal endothelial cells resulted in detrimental plasmin activation that compromised liver vascular integrity and contributed to embryonic lethality. To validate these data, we performed in situ zymography on fresh liver sections from E12.5 control and mutant embryos 2 days prior to the hemorrhage and necrosis we observed histologically in Chd4fl/fl Lyve1-Cre+ livers. In situ zymography revealed elevated plasmin activity in a strikingly consistent pattern at the periphery of Chd4fl/fl Lyve1-Cre+ left liver lobes, whereas plasmin activity was not detectable in livers from control littermates (Figure 6, A and B). Although Chd4fl/fl Lyve1-Cre+ livers displayed no grossly visible hemorrhage at E12.5 (Figure 6D), bleeding was consistently obvious in the periphery of the left lobe in E13.5 Chd4fl/fl Lyve1-Cre+ livers (6 of 6 livers; Figure 6F). Therefore, plasmin elevation and hemorrhage occurred in corresponding regions of Chd4fl/fl Lyve1-Cre+ livers prior to embryonic lethality. Moreover, TUNEL staining indicated that cell death also initiated at the periphery of E13.5 Chd4fl/fl Lyve1-Cre+ left liver lobes and progressed throughout the remaining lobes by E14.5 (Supplemental Figure 8, B and F). Genetic uPA reduction diminished plasmin activity in Chd4fl/fl Lyve1-Cre+ livers, as expected, but did not rescue hepatic plasmin activity or cell death to levels seen in littermate control livers (Supplemental Figure 8). This residual plasmin activity and TUNEL staining corresponds with the hemorrhage and parenchymal breakdown that is histologically detectable in uPA+/– Chd4fl/fl Lyve1-Cre+ and uPA–/– Chd4fl/fl Lyve1-Cre+ livers at E14.5 (Figure 5, G–L).
 Figure 6
Figure 6Plasmin activity is increased prior to hemorrhage in Chd4fl/fl Lyve1-Cre+ livers. (A and B) Unfixed sections from E12.5 control (A) and Chd4fl/fl Lyve1-Cre+ (B) embryos were subjected to in situ zymography for detection of plasmin activity. After the addition of exogenous plasminogen, plasmin activity (green fluorescence; white arrows) was substantially higher in the left lobe from Chd4fl/fl Lyve1-Cre+ livers than in comparable regions from control livers. The 5× magnified insets correspond to the areas marked with white asterisks. n = 4 embryos per genotype. (C–F) Livers were dissected from E12.5 (C and D) and E13.5 (E and F) littermate control and Chd4fl/fl Lyve1-Cre+ embryos and photographed. Hemorrhage (white arrows) was grossly evident on the periphery of the left lobe of all 6 mutant livers analyzed at E13.5 (F). n = 3 embryos per genotype for C and D. n = 6 embryos per genotype for E and F. Scale bars: 500 μm. PC, pericardial cavity; St, stomach; ML, medial lobe; RL, right lobe; LL, left lobe; CL, caudate lobe.
Finally, to determine whether elevated plasmin activity correlated with extracellular matrix degradation that could account for vascular fragility in Chd4fl/fl Lyve1-Cre+ livers, we analyzed lysates generated from the left lobes of E13.5 control and mutant livers by immunoblotting. We saw significant reduction of the vascular matrix component laminin, which can be directly cleaved by plasmin (29), in Chd4fl/fl Lyve1-Cre+ livers and a modest reduction of the matrix components fibronectin and collagen IV (Figure 7, A and B). By contrast, fibrinogen levels were not reduced in Chd4fl/fl Lyve1-Cre+ livers (Figure 7, A and B). In addition, we found that CHD4 was significantly reduced in lysates from E13.5 Chd4fl/fl Lyve1-Cre+ versus control left liver lobes, indicating that at least half of the CHD4 in the developing left lobe is expressed in LYVE1+ cells (Figure 7, C and D). Moreover, CHD4 expression was significantly higher in the left liver lobe than in the remaining lobes at E13.5 and was dramatically reduced throughout the liver by E19.5 (Figure 7, E and F). Collectively, our data indicate that in addition to protecting embryonic LV thrombi from plasmin-mediated fibrinolysis and dissolution, CHD4 plays an important role in protecting developing hepatic sinusoidal blood vessels — particularly in the left lobe — from excessive plasmin activation and extracellular matrix degradation (Figure 8).
 Figure 7
Figure 7The extracellular matrix component laminin is diminished in Chd4fl/fl Lyve1-Cre+ left liver lobes. (A) Expression of fibrinogen or of the extracellular matrix components laminin, fibronectin, and collagen IV were assessed by immunoblotting lysates generated from the left lobes of E13.5 control and Chd4fl/fl Lyve1-Cre+ livers. (B) Laminin, which is directly cleaved by plasmin, was significantly reduced in Chd4fl/fl Lyve1-Cre+ livers compared with controls. n = 3 embryos per genotype; experiments were repeated twice. Values expressed as relative normalized expression are mean ± SEM. P = 0.0002 as calculated by Student’s t test. (C) Expression of CHD4 and LYVE1 was assessed by immunoblotting lysates generated from the left lobes of E13.5 control and Chd4fl/fl Lyve1-Cre+ livers. (D) CHD4 expression was significantly reduced in Chd4fl/fl Lyve1-Cre+ left liver lobe samples compared with controls, while LYVE1 expression remained unchanged. n = 3 embryos per genotype; experiments were repeated twice. Values expressed as relative normalized expression are mean ± SEM. P = 0.043 as calculated by Student’s t test. (E) Expression of CHD4 was analyzed in liver samples taken from the left lobe and remaining lobes of E13.5 and E19.5 control embryos. Lanes were run on the same gel but were noncontiguous. (F) CHD4 expression was higher in the left lobe than in the remaining lobes of E13.5 livers (P = 0.007), but CHD4 expression was dramatically decreased throughout the liver by E19.5 (P = 0.002 for left lobe and P = 0.004 for remaining lobes). n = 3 embryos per genotype; experiments were repeated twice. Values expressed as relative normalized expression are mean ± SEM. P values were calculated by one-way ANOVA using Holm-Šidák’s multiple comparisons test.
 Figure 8
Figure 8Effects of plasmin activation in embryonic Chd4fl/fl Lyve1-Cre+ lymphatic and liver vasculature. Transcriptional upregulation of uPAR in Chd4-deficient LYVE1+ LECs and liver sinusoidal endothelial cells leads to elevated plasmin activity and two different phenotypes. In the lymphatic system, elevated plasmin increases fibrinolysis of the lymphovenous valve (LVV) thrombi that prevent blood from entering the lymphatic vasculature. In the liver, elevated plasmin causes degradation of the extracellular matrix (ECM) protein laminin around sinusoidal blood vessels, which leads to vascular rupture and embryonic lethality.
-
Discussion
A sophisticated network of protein activators and inhibitors regulates the serine protease plasmin, which can degrade many extracellular proteins, including fibrin (23, 30). In this article we show that Chd4 deletion by Lyve1-Cre results in elevated plasmin activity through increased transcription of the urokinase receptor uPAR in LYVE1+ cells, which triggers fibrinolysis of thrombi at the LV valves. The dissolution of the thrombi impairs LV hemostasis and facilitates the entry of blood into the lymphatic circulation, despite the presence of morphologically normal LV valves in Chd4fl/fl Lyve1-Cre+ mutants. We also show that Chd4 deletion with Lyve1-Cre results in rupture of liver sinusoids due to excessive plasmin activation and vascular extracellular matrix degradation. Genetic reduction of uPA ameliorated both the dissolution of LV thrombi and the liver hemorrhage in Chd4fl/fl Lyve1-Cre+ mutants, indicating that the regulation of plasmin activation by CHD4 has important functions in both lymphatic and hepatic blood vasculature during embryonic development. However, we still detected some plasmin activation and hemorrhage in uPA–/– Chd4fl/fl Lyve1-Cre+ livers, which likely explains why those embryos eventually died. Since tPA is an important alternative plasmin activator to uPA, we assessed tPA transcripts in LYVE1+ cells isolated from Chd4fl/fl Lyve1-Cre+ embryos but saw no aberrant upregulation at E13.5 (Figure 3E). This finding suggests that CHD4 has additional indirect or alternative means of regulating plasmin activation during embryonic development. We suspect that Chd4fl/fl Lyve1-Cre+ embryos on a plasminogen-deficient background would show complete resolution of embryonic blood-filled lymphatics and liver hemorrhage. Further studies will also be required to eliminate a role for Chd4fl/fl Lyve1-Cre+ macrophages in contributing to the lymphatic and liver phenotypes we observed, since a subset of macrophages also express LYVE1 during embryonic development (31).
Since LV thrombi persist after birth (5), it is possible that they remain susceptible to plasmin-mediated degradation. This is an important question, because fibrinolytic drugs, including recombinant tPA, are administered to patients to activate plasmin and dissolve life-threatening blood clots (32). Our study does not address the capacity for CHD4 to suppress plasmin activation in postnatal lymphatics or LV valves. It is possible that CHD4 becomes less important for regulating plasmin activation on LECs after birth as the plasminogen activator inhibitor (PAI) proteins PAI-1 and PAI-2 become more widely expressed and functional. The insignificance of embryonic PAI activity is demonstrated by the lack of developmental defects in PAI-deficient embryos and by the limited expression of PAI-2 at E15.5 (33–35). Genetic studies will provide the best opportunity for deciphering the relative impact of CHD4 and the PAIs on LV thrombus protection after birth. Such studies will also clarify whether extracellular matrix at the LV valves is susceptible to plasmin-mediated degradation that adversely impacts LV valve function in postnatal animals. Undifferentiated mesenchymal precursor cells are first detectable at the LV valves in E14.5 embryos (4). Since we already detected blood-filled lymphatics and edema in Chd4fl/fl Lyve1-Cre+ embryos by E14.5 (Figure 1), we suspect that the lymphatic phenotypes we observed initiate prior to the time at which extracellular matrix is secreted or functional at the LV valves. In support of this hypothesis, we did not detect robust expression of matrix markers in E13.5 LV valves from control embryos (Supplemental Figure 9). However, by E18.5 mesenchymal cells at the LV valves display more mature mural cell markers and enlarged secretory endoplasmic reticulum, indicating that they are capable of secreting matrix components (4). Therefore, postnatal LV valves may require matrix for their function, and this matrix — like the LV thrombi — may be degraded by excessive plasmin activity.
The susceptibility of embryonic hepatic sinusoids to plasmin-mediated degradation also raises important questions about the impact of plasmin on postnatal liver vessels. Laminin, which can be directly cleaved by plasmin (29), is highly expressed around embryonic sinusoids but is downregulated in perisinusoidal spaces close to birth (36). Interestingly, laminin is transiently upregulated again around sinusoidal vessels during liver regeneration (37). These expression patterns suggest that laminin plays an important role in liver sinusoidal development throughout life. Since our current study indicates that plasmin-mediated laminin degradation correlates with embryonic liver hemorrhage, we question whether laminin-rich sinusoidal vessels in regenerating livers are likewise susceptible to plasmin-mediated degradation and rupture. We also question whether CHD4 expression, which we found increasingly downregulated in the liver close to birth (Figure 7, E and F), is elevated again — like laminin — in regenerating livers. Such an expression pattern might indicate that CHD4 recapitulates its embryonic role of protecting laminin-rich sinusoids from plasmin-mediated rupture during postnatal liver regeneration. Alternatively, the PAIs may be sufficient to fulfill this role after birth.
One type of liver injury that is characterized by excessive plasmin activity and vascular rupture is induced by acetaminophen (N-acetyl-p-aminophenol; APAP) overdose. Pharmacological inhibition or genetic depletion of the plasmin zymogen plasminogen significantly reduces APAP-induced liver damage in mice (38). Conversely, hepatic fibrosis, necrosis, and hemorrhage are enhanced in APAP-treated mice with a global PAI-1 deficiency (38). Interestingly, genetic or enzymatic depletion of fibrinogen fails to ameliorate APAP-induced liver injury (38), indicating that plasmin-mediated activities other than fibrinolysis contribute to murine APAP-induced liver damage. Based on our new results in Chd4fl/fl Lyve1-Cre+ embryos — which have liver hemorrhage and hepatic cell death similar to those seen with APAP-induced injury — we propose that plasmin-mediated degradation of laminin around hepatic blood vessels may contribute to the pathology of APAP-induced liver injury. If this is true, acute treatment with plasmin inhibitors following APAP overdose could protect laminin-rich hepatic vessels from rupture.
If plasmin-mediated laminin degradation weakens hepatic sinusoidal vessels, as our embryonic data suggest, those vessels located at sites of high hemodynamic stress might be most susceptible to rupture. During embryogenesis, half of the circulating blood enters the developing liver, while the other half bypasses the liver and is shunted toward the heart through the ductus venosus, a transient blood vessel that disappears after birth (39). Of the blood entering the embryonic liver, 80% arrives through the umbilical vein, which transports oxygenated blood from the placenta and delivers it directly to the developing sinusoidal network (39). The hemodynamic forces imparted as blood flows through the sinusoids are believed to organize hepatocytes into linear plates, through a process that begins in the left lobe and progressively occurs in the right lobe (39). This progression is thought to reflect the fact that the developing left lobe receives all its blood from the umbilical vein, whereas the right lobe receives blood from both the umbilical and the portal veins (40). Therefore, hemodynamic forces are likely higher in the developing left lobe than the right lobe. Our detection of plasmin activity and subsequent hemorrhage originating in the left lobe of Chd4fl/fl Lyve1-Cre+ livers indicates that sinusoids weakened by matrix degradation are most susceptible to rupture under hemodynamic stress. We propose that CHD4 expression is greatest in the developing left lobe to protect the hepatic sinusoids under the highest hemodynamic stress from plasmin-mediated laminin degradation and rupture.
Finally, it is worth noting that the liver is the key site of synthesis of most proteins required for the regulation of coagulation and fibrinolysis. Because hepatocytes and sinusoidal endothelial cells produce clotting and inhibitory factors that circulate in the bloodstream (41, 42), liver injury and disease result in diminished production of these factors and subsequently a variety of coagulation disorders (7, 43). Therefore, the plasmin-induced liver damage we detected in Chd4fl/fl Lyve1-Cre+ livers as early as E13.5 may contribute to the deficit of fibrin-rich LV thrombi we detected in those embryos. If clotting factors are reduced in Chd4fl/fl Lyve1-Cre+ embryos due to hepatic necrosis, thrombin generation and fibrinogen cleavage on platelets that are activated by PDPN/CLEC-2 signaling at the LV valves may be compromised. Further investigation will be required to determine whether genetic uPA deletion rescues LV thrombi in Chd4fl/fl Lyve1-Cre+ embryos by diminishing plasmin-mediated fibrinolysis of the thrombi or by reducing plasmin-mediated liver damage. Meanwhile, it is worthwhile considering the connection among plasmin, liver damage, and lymphatic dysfunction in clinical contexts. For example, impaired formation or excessive dissolution of LV thrombi may contribute to the cerebral edema that correlates with APAP-induced liver damage (44). Likewise, peliosis hepatis is a rare liver disease with unknown causes and with phenotypes resembling those seen in Chd4fl/fl Lyve1-Cre+ embryos, including hepatic lesions and sinusoidal dilation that can progress to spontaneous rupture, hemorrhage, and death (45). Lymphatic dysfunction characterized by tissue edema is also observed in rodents and patients with peliosis hepatis (46–48). In light of our current study, we propose that excessive plasmin activity should be investigated as a pathogenic mediator of this and other diseases characterized by hepatic sinusoidal rupture and lymphedema.
-
Methods
Mice. Chd4-floxed mice (Chd4fl/fl), uPA+/− mice (The Jackson Laboratory, 002509), VE-cadherin-Cre transgenic mice (The Jackson Laboratory, 006137), Lyve1-Cre knock-in mice (The Jackson Laboratory, 012601), PF4-Cre transgenic mice (The Jackson Laboratory, 008535), and ROSAmT/mG knock-in mice (The Jackson Laboratory, 007676) have been described (12, 14, 22, 49–51). Chd4-floxed mice were a gift from Katia Georgopoulos (Massachusetts General Hospital, Charlestown, Massachusetts, USA); uPA+/– mice were a gift from Li Zhang (University of Maryland School of Medicine, Baltimore, Maryland, USA); VE-cadherin-Cre transgenic mice were a gift from Luisa Iruela-Arispe (University of California, Los Angeles, California, USA). All mice were maintained on a mixed genetic background at the Oklahoma Medical Research Foundation animal facility. Chd4-floxed and uPA-deficient embryos and mice were genotyped by PCR, as described previously (9). The VE-cadherin-Cre transgene was detected with a forward primer (5′-GCAGGCAGCTCACAAAGGAACAAT-3′) and a reverse primer (5′-ATCACTCGTTGCATCGACCGGTAA-3′) that yielded a 300-bp amplicon. The WT Lyve1 allele was amplified with a forward primer (5′-TGCCACCTGAAGTCTCTCCT-3′) and a reverse primer (5′-TGAGCCACAGAAGGGTTAGG-3′) that yielded a 425-bp amplicon. The targeted Lyve1 allele was amplified by forward primer (5′-GAGGATGGGGACTGAAACTG-3′) and the same reverse primer used for detecting the WT allele to yield a 210-bp amplicon. The PF4-Cre transgene was detected with a forward primer (5′-CCCATACAGCACACCTTTTG-3′) and a reverse primer (5′-TGCACAGTCAGCAGGTT-3′) that yielded a 450-bp amplicon. The WT ROSA allele was detected with a forward primer (5′-CTCTGCTGCCTCCTGGCTTCT-3′) and a reverse primer (5′-CGAGGCGGATCACAAGCAATA-3′) that yielded a 330-bp amplicon. The targeted ROSAmT/mG allele was amplified by a reverse primer (5′-TCAATGGGCGGGGGTCGTT-3′) and the same ROSA forward primer to yield a 250-bp amplicon.
Histological staining. H&E staining was performed on paraffin sections, as described previously (52), with slight modification. Dissected embryos (E12.5–E15.5) were fixed in 4% paraformaldehyde for 48 hours and then immersed in 70% ethanol for 24 hours prior to sample embedding. Using a Microm 355 S microtome (Microm International), sections were cut at 8 μm thickness, mounted on HistoBond microscope slides (VWR), and allowed to dry overnight prior to staining.
Immunofluorescence. Chd4fl/fl Lyve1-Cre+, uPA+/– Chd4fl/fl Lyve1-Cre+, uPA–/– Chd4fl/fl Lyve1-Cre+, or littermate control embryos were dissected, embedded in paraffin, and sectioned as stated above. Yolk sac tissue collected prior to embedding was used for genotyping. Sections were deparaffinized using xylene and rehydrated through a decreasing alcohol gradient. Antigen retrieval was then performed for 20 minutes using Antigen Unmasking Solution (Vector Laboratories) and a vegetable steamer (West Bend). Sections were blocked in blocking solution (5% normal goat serum/5% normal donkey serum/0.3% BSA/0.1% Triton X-100 in PBS) for 1 hour at room temperature. Sections were incubated in primary antibody at concentrations of 1:20 to 1:200 in blocking solution overnight at 4°C, washed three times (3 minutes each) with PBS, then incubated for 1 hour at room temperature in secondary antibody at a 1:500 concentration in blocking solution along with 10 μg/ml Hoechst stain (VWR 89139-124). Sections were then washed three times (3 minutes each) with PBS and coverslipped with 2.5% DABCO/90% glycerol/PBS, pH 8.6. Primary antibodies used for immunohistochemistry were rabbit anti-fibrin(ogen) (1:20, Santa Cruz Biotechnology Inc., 33582), biotinylated goat anti-LYVE1 (1:50, R&D Systems, BAF2125), rat anti-TER119 (1:20, BD Biosciences, 550565), goat anti-PROX1 (1:50, R&D Systems, AF2727), rat anti-CD41 (1:50, BD Biosciences, 553847), rabbit anti-CHD4 (1:100, Active Motif, 39289), and Syrian hamster anti-PDPN (1:200, clone 8.1.1, Developmental Studies Hybridoma Bank). Secondary antibodies used were streptavidin–Alexa Fluor 488, streptavadin–Alexa Fluor 594, and Alexa Fluor 488–goat anti-rabbit IgG; Cy3 anti–Syrian hamster IgG (Jackson ImmunoResearch Laboratories Inc.); and Alexa Fluor 594–goat anti-rabbit IgG (Invitrogen).
Primary LYVE1+ cell isolation. E13.5 embryos were digested with collagenase-DNase solution (1.5 mg/ml collagenase, 25 mg/ml DNase, 25 mM HEPES in DMEM) for 30 minutes at 37°C. Cells were washed once with PBS/0.1% BSA and resuspended in DMEM. Dynabeads (Life Technologies) conjugated to LYVE1 antibody (R&D Systems, AF2125) were added, and samples were incubated for 30 minutes at 4°C with rotation. Immunoprecipitated cells were washed with PBS/0.1% BSA and eluted in TRIzol (Life Technologies).
qPCR. To analyze transcript levels, we purified total RNA isolated from primary LYVE1+ cells using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions. cDNA was prepared using the iScript Advanced cDNA Synthesis Kit (Bio-Rad, 172-5037), and qPCR was performed using 2× SYBR green qPCR master mix (Life Technologies) and the CFX96 Real-Time System (Bio-Rad) with gene-specific primers. The following mouse-specific primers were used for qPCR: Chd4 (5′-TCCTCTGTCCACCATCATCA-3′ and 5′-ACCCAAGATGGCCATATCAA-3′); Pdpn (5′-ATGGCTTGCCAGTAGTCACC-3′ and 5′-AGCTCTTTAGGGCGAGAACC-3′); uPA (5′-GCGCCTTGGTGGTGAAAAAC-3′ and 5′-TTGTAGGACACGCATACACCT-3′); uPAR (5′-GGCTTAGATGTGCTGGGAAA-3′ and 5′-CAATGAGGCTGAGTTGAGCA-3′); Thbs1 (5′-CCAAAGCCTGCAAGAAAGAC-3′ and 5′-CCTGCTTGTTGCAAACTTGA-3′); tPA (5′-CTGAGGTCACAGTCCAAGCA-3′ and 5′-TCAGCCGGTCAGAGAAGAAT-3′); Gapdh (5′-TCAACGGCACAGTCAAGG-3′ and 5′-ACTCCACGACATACTCAGC-3′); and Actb (5′-TGTTACCAACTGGGACGACA-3′ and 5′-GGGGTGTTGAAGGTCTCAAA-3′).
qPCR analysis. The relative fold change in transcription was determined using the comparative CT method and the housekeeping genes Gapdh and Actb as internal controls. Data from at least 4 independent sets of littermate control and mutant embryos were represented in a scatter plot. Error bars indicate SEM.
In situ zymography. E12.5, E13.5, and E14.5 embryos were cryosectioned without prior fixation. Sections (16 μm) of control, Chd4fl/fl Lyve1-Cre+, uPA+/– Chd4fl/fl Lyve1-Cre+, and uPA–/– Chd4fl/fl Lyve1-Cre+ embryos were affixed to slides and were overlaid with in situ zymography solution (ISZS) consisting of 1% low-melting-point agarose (Invitrogen), 0.1 mg/ml quenched BODIPY FL casein from the EnzChek Protease Assay Kit (Invitrogen), 1 U/ml human Glu-plasminogen (Sigma-Aldrich), and 10 μg/ml Hoechst (VWR 89139-124). Overlaid sections were coverslipped and incubated for 1 hour at 37°C. Casein cleavage was detected by fluorescence microscopy. Plasminogen-free ISZS was used as a negative control to verify that casein cleavage was plasmin dependent.
In situ cell death detection. After 48 hours fixation in 4% paraformaldehyde, E12.5–E14.5 control, Chd4fl/fl Lyve1-Cre+, uPA+/– Chd4fl/fl Lyve1-Cre+, and uPA–/– Chd4fl/fl Lyve1-Cre+ embryos were embedded in paraffin and sectioned at 10 μm for TUNEL staining (Roche In Situ Cell Death Detection, Fluorescein, 11684795910). In brief, sections were deparaffinized in xylene, rehydrated, subjected to proteinase-K digestion (20 μg/ml in 10 μM Tris-HCl pH 7.4) for 30 minutes at 37°C, and rinsed twice with PBS. Sections were then treated with TUNEL reaction mixture (fluorescein-dUTP) for 1 hour at 37°C and rinsed twice with PBS. Hoechst was applied for 5 minutes before slides were coverslipped with 2.5% DABCO/90% glycerol/PBS, pH 8.6. TUNEL was detected by fluorescence microscopy. TUNEL reaction mixture–free sections were used as a negative control.
Fluorescence microscopy and image acquisition. Gross embryonic images were obtained with a Nikon SMZ800 stereomicroscope and Nikon DS-Fi1 camera and monitor. Brightfield histological images were obtained with a Nikon Eclipse 80i microscope using 10× (NA 0.3), 20× (NA 0.5), and 40× (NA 0.75) objectives and a Nikon DS-Fi1 camera. Fluorescence images were obtained with a Nikon Eclipse 80i microscope using 10× (NA 0.3), 20× (NA 0.5), and 40× (NA 0.75) objectives, an X-cite 120Q light source, and a Nikon DS-Qi1Mc camera. NIS-Elements AR3.0 (Nikon) software was used for all brightfield and fluorescence image acquisition and assembly.
Immunoblotting. E13.5 left liver lobes and the remaining liver lobes (medial, right, and caudate) were dissected and placed in RIPA buffer (10 mM Tris, 1 mM EDTA, 1% Triton X-100, 1.1% sodium deoxycholate, 0.1% SDS, and 140 mM NaCl) with protease inhibitor cocktail (Invitrogen, P8340) and Dounce homogenized on ice. Lysates were centrifuged at 9,300 g for 15 minutes at 4°C. The supernatant was collected, and protein concentration was determined using the Pierce BCA Protein Assay Kit (Thermo Scientific, 23227) and a NanoDrop 2000 (Thermo Scientific). Lysates were diluted in 2× Laemmli buffer, and 15 μg protein was loaded in each well of an SDS-PAGE gel (9%) and a Mini Trans-Blot Cell setup (Bio-Rad), electrophoresed, then transferred to a PVDF membrane that was blocked in 5% milk-TBST for 45 minutes. Primary antibodies (diluted in 5% milk-TBST) were incubated at 4°C overnight with gentle agitation, then membranes were washed two times (5 minutes each) in TBST. HRP-conjugated secondary antibodies (diluted in 5% milk-TBST) were applied at room temperature for 1 hour with gentle agitation, and membranes were then washed three times (10 minutes each) in TBST. Secondary antibodies were detected through autoradiography using enhanced chemiluminescence (ECL, GE Healthcare, RPN2209), a Konica Minolta processor (SRX-101A), and x-ray film (Thermo Scientific, 34090). Rabbit anti-GAPDH (1:10,000, Sigma-Aldrich, G9545) was used as a loading control. Densities for rabbit anti-laminin (1:1,000, Abcam, 11575), rabbit anti–collagen IV (1:500, Abcam, 19808), mouse anti-fibronectin (1:500, Sigma-Aldrich, F1640), rabbit anti-fibrinogen (1:20, Santa Cruz Biotechnology Inc., 33582), rabbit anti-CHD4 (1:500, Abcam, 72418), and goat anti-LYVE1 (1:500, R&D Systems, AF2125) were normalized to GAPDH using ImageJ software (1.46r, https://imagej.nih.gov/ij/).
Statistics. χ2 analysis was performed based on the observed genotypes produced from Chd4fl/fl × Chd4fl/+ VE-cadherin-Cre+ crosses and from Chd4fl/fl × Chd4fl/+ Lyve1-Cre+ crosses. Because 4 categories of genotypes were expected from each of these crosses, 3 degrees of freedom (dof) were used for the analyses. For qPCR and immunoblotting, statistical significance was determined based on a two-tailed Student’s t test or a one-way ANOVA using Holm-Šidák’s multiple comparisons test (GraphPad Prism 6.0). Error bars represent the mean ± SEM. P values of less than or equal to 0.05 were considered statistically significant for all analyses.
Study approval. The Oklahoma Medical Research Foundation (OMRF) is accredited by Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) International and follows the Public Health Service Policy for the Care and Use of Laboratory Animals. Animal care was provided in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals (National Academies Press. 2011). The OMRF Institutional Animal Care and Use Committee approved all animal use protocols.
-
Author contributions
PLC, CDC, and CTG designed the research studies. PLC, JJP, CDC, and SG conducted the experiments. PLC, JJP, CDC, LX, RSS, and CTG interpreted the data. LX, RSS, and CTG provided reagents. PLC and CTG wrote the manuscript. CDC, LX, and RSS provided editorial advice.
-
Acknowledgments
We thank Vijay Muthukumar and Eileen Dilks for technical assistance with these studies. We also thank Mark Kahn and members of the Griffin laboratory for valuable advice and feedback on this manuscript. This work was supported by NIH grants HL111178 to C.T. Griffin and HD083418 and HL085607 to L. Xia, and American Heart Association Postdoctoral Fellowship 15POST21180005 to P.L. Crosswhite.
Address correspondence to: Courtney T. Griffin, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma 73104, USA. Phone: 405.271.7073; E-mail: courtney-griffin@omrf.org.
-
Footnotes
Carol D. Curtis’s present address is: i2E, Inc., Oklahoma City, Oklahoma, USA.
Joanna J. Podsiadlowska’s present address is: University of Warsaw, Warsaw, Poland.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2016;126(6):2254–2266. doi:10.1172/JCI84652.
-
References
- Wiig H, Swartz MA. Interstitial fluid and lymph formation and transport: physiological regulation and roles in inflammation and cancer. Physiol Rev. 2012;92(3):1005–1060.
- Srinivasan RS, Oliver G. Prox1 dosage controls the number of lymphatic endothelial cell progenitors and the formation of the lymphovenous valves. Genes Dev. 2011;25(20):2187–2197.
- Turner CJ, Badu-Nkansah K, Crowley D, van der Flier A, Hynes RO. Integrin-alpha5beta1 is not required for mural cell functions during development of blood vessels but is required for lymphatic-blood vessel separation and lymphovenous valve formation. Dev Biol. 2014;392(2):381–392.
- Geng X, et al. Multiple mouse models of primary lymphedema exhibit distinct defects in lymphovenous valve development. Dev Biol. 2016;409(1):218–233.
- Hess PR, et al. Platelets mediate lymphovenous hemostasis to maintain blood-lymphatic separation throughout life. J Clin Invest. 2014;124(1):273–284.
- Suzuki-Inoue K, Inoue O, Ozaki Y. Novel platelet activation receptor CLEC-2: from discovery to prospects. J Thromb Haemost. 2011;9 Suppl 1:44–55.View this article via: PubMed Google Scholar
- Versteeg HH, Heemskerk JW, Levi M, Reitsma PH. New fundamentals in hemostasis. Physiol Rev. 2013;93(1):327–358.
- Cesarman-Maus G, Hajjar KA. Molecular mechanisms of fibrinolysis. Br J Haematol. 2005;129(3):307–321.
- Ingram KG, Curtis CD, Silasi-Mansat R, Lupu F, Griffin CT. The NuRD chromatin-remodeling enzyme CHD4 promotes embryonic vascular integrity by transcriptionally regulating extracellular matrix proteolysis. PLoS Genet. 2013;9(12):e1004031.
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304.
- Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26(37):5433–5438.
- Alva JA, et al. VE-Cadherin-Cre-recombinase transgenic mouse: a tool for lineage analysis and gene deletion in endothelial cells. Dev Dyn. 2006;235(3):759–767.
- Schulte-Merker S, Sabine A, Petrova TV. Lymphatic vascular morphogenesis in development, physiology, and disease. J Cell Biol. 2011;193(4):607–618.
- Pham TH, et al. Lymphatic endothelial cell sphingosine kinase activity is required for lymphocyte egress and lymphatic patterning. J Exp Med. 2010;207(1):17–27.
- Fu J, et al. Endothelial cell O-glycan deficiency causes blood/lymphatic misconnections and consequent fatty liver disease in mice. J Clin Invest. 2008;118(11):3725–3737.
- Bertozzi CC, et al. Platelets regulate lymphatic vascular development through CLEC-2-SLP-76 signaling. Blood. 2010;116(4):661–670.
- Suzuki-Inoue K, et al. Essential in vivo roles of the C-type lectin receptor CLEC-2: embryonic/neonatal lethality of CLEC-2-deficient mice by blood/lymphatic misconnections and impaired thrombus formation of CLEC-2-deficient platelets. J Biol Chem. 2010;285(32):24494–24507.
- Abtahian F, et al. Regulation of blood and lymphatic vascular separation by signaling proteins SLP-76 and Syk. Science. 2003;299(5604):247–251.
- Alitalo K, Tammela T, Petrova TV. Lymphangiogenesis in development and human disease. Nature. 2005;438(7070):946–953.
- Uhrin P, et al. Novel function for blood platelets and podoplanin in developmental separation of blood and lymphatic circulation. Blood. 2010;115(19):3997–4005.
- Srinivasan RS, et al. The nuclear hormone receptor Coup-TFII is required for the initiation and early maintenance of Prox1 expression in lymphatic endothelial cells. Genes Dev. 2010;24(7):696–707.
- Tiedt R, Schomber T, Hao-Shen H, Skoda RC. Pf4-Cre transgenic mice allow the generation of lineage-restricted gene knockouts for studying megakaryocyte and platelet function in vivo. Blood. 2007;109(4):1503–1506.
- Collen D. The plasminogen (fibrinolytic) system. Thromb Haemost. 1999;82(2):259–270.View this article via: PubMed Google Scholar
- Alfano D, et al. The urokinase plasminogen activator and its receptor: role in cell growth and apoptosis. Thromb Haemost. 2005;93(2):205–211.View this article via: PubMed Google Scholar
- Carriero MV, Stoppelli MP. The urokinase-type plasminogen activator and the generation of inhibitors of urokinase activity and signaling. Curr Pharm Des. 2011;17(19):1944–1961.
- Dellinger MT, Meadows SM, Wynne K, Cleaver O, Brekken RA. Vascular endothelial growth factor receptor-2 promotes the development of the lymphatic vasculature. PLoS One. 2013;8(9):e74686.
- Gordon EJ, Gale NW, Harvey NL. Expression of the hyaluronan receptor LYVE-1 is not restricted to the lymphatic vasculature; LYVE-1 is also expressed on embryonic blood vessels. Dev Dyn. 2008;237(7):1901–1909.View this article via: PubMed Google Scholar
- Mouta Carreira C, et al. LYVE-1 is not restricted to the lymph vessels: expression in normal liver blood sinusoids and down-regulation in human liver cancer and cirrhosis. Cancer Res. 2001;61(22):8079–8084.View this article via: PubMed Google Scholar
- Liotta LA, Goldfarb RH, Terranova VP. Cleavage of laminin by thrombin and plasmin: alpha thrombin selectively cleaves the beta chain of laminin. Thromb Res. 1981;21(6):663–673.
- Pepper MS. Role of the matrix metalloproteinase and plasminogen activator-plasmin systems in angiogenesis. Arterioscler Thromb Vasc Biol. 2001;21(7):1104–1117.
- Gordon EJ, Rao S, Pollard JW, Nutt SL, Lang RA, Harvey NL. Macrophages define dermal lymphatic vessel calibre during development by regulating lymphatic endothelial cell proliferation. Development. 2010;137(22):3899–3910.
- Wardlaw JM, et al. Recombinant tissue plasminogen activator for acute ischaemic stroke: an updated systematic review and meta-analysis. Lancet. 2012;379(9834):2364–2372.
- Carmeliet P, et al. Plasminogen activator inhibitor-1 gene-deficient mice. II. Effects on hemostasis, thrombosis, and thrombolysis. J Clin Invest. 1993;92(6):2756–2760.
- Dougherty KM, Pearson JM, Yang AY, Westrick RJ, Baker MS, Ginsburg D. The plasminogen activator inhibitor-2 gene is not required for normal murine development or survival. Proc Natl Acad Sci U S A. 1999;96(2):686–691.
- Kawata Y, Mimuro J, Kaneko M, Shimada K, Sakata Y. Expression of plasminogen activator inhibitor 2 in the adult and embryonic mouse tissues. Thromb Haemost. 1996;76(4):569–576.View this article via: PubMed Google Scholar
- Amenta PS, Harrison D. Expression and potential role of the extracellular matrix in hepatic ontogenesis: a review. Microsc Res Tech. 1997;39(4):372–386.
- Wewer UM, Engvall E, Paulsson M, Yamada Y, Albrechtsen R. Laminin A, B1, B2, S and M subunits in the postnatal rat liver development and after partial hepatectomy. Lab Invest. 1992;66(3):378–389.View this article via: PubMed Google Scholar
- Sullivan BP, Kassel KM, Jone A, Flick MJ, Luyendyk JP. Fibrin(ogen)-independent role of plasminogen activators in acetaminophen-induced liver injury. Am J Pathol. 2012;180(6):2321–2329.
- Collardeau-Frachon S, Scoazec JY. Vascular development and differentiation during human liver organogenesis. Anat Rec (Hoboken). 2008;291(6):614–627.
- Haugen G, Kiserud T, Godfrey K, Crozier S, Hanson M. Portal and umbilical venous blood supply to the liver in the human fetus near term. Ultrasound Obstet Gynecol. 2004;24(6):599–605.
- Everett LA, Cleuren AC, Khoriaty RN, Ginsburg D. Murine coagulation factor VIII is synthesized in endothelial cells. Blood. 2014;123(24):3697–3705.
- DeSancho MT, Pastores SM. , The liver and coagulation. Rodés J, Benhamou J-P, Blei A, Reichen J, Rizzetto M, eds. Textbook of Hepatology: From Basic Science to Clinical Practice.
- Amitrano L, Guardascione MA, Brancaccio V, Balzano A. Coagulation disorders in liver disease. Semin Liver Dis. 2002;22(1):83–96.
- Makin AJ, Wendon J, Williams R. A 7-year experience of severe acetaminophen-induced hepatotoxicity (1987-1993). Gastroenterology. 1995;109(6):1907–1916.
- Yanoff M, Rawson AJ. Peliosis hepatis. An anatomic study with demonstration of two varieties. Arch Pathol. 1964;77:159–165.View this article via: PubMed Google Scholar
- Corpa MV, Bacchi MM, Bacchi CE, Coelho KI. Peliosis hepatis associated with lymphoplasmacytic lymphoma: an autopsy case report. Arch Pathol Lab Med. 2004;128(11):1283–1285.View this article via: PubMed Google Scholar
- Lee KP. Peliosis hepatis-like lesion in aging rats. Vet Pathol. 1983;20(4):410–423.
- Yoshioka K, et al. Peliosis hepatis associated with idiopathic restrictive cardiomyopathy. Intern Med. 1998;37(1):51–55.
- Williams CJ, et al. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20(6):719–733.
- Carmeliet P, et al. Physiological consequences of loss of plasminogen activator gene function in mice. Nature. 1994;368(6470):419–424.
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605.
- Griffin CT, Brennan J, Magnuson T. The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development. 2008;135(3):493–500.
-
Version history
- Version 1 (May 3, 2016): No description
- Version 2 (June 1, 2016): No description



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)











