Advertisement
Research Article Free access | 10.1172/JCI41402
Mitigation of hematologic radiation toxicity in mice through pharmacological quiescence induced by CDK4/6 inhibition
Søren M. Johnson,1,2 Chad D. Torrice,1,2 Jessica F. Bell,1,3 Kimberly B. Monahan,1,2 Qi Jiang,4 Yong Wang,5 Matthew R. Ramsey,1,2 Jian Jin,6 Kwok-Kin Wong,7 Lishan Su,4 Daohong Zhou,5 and Norman E. Sharpless1,2,8
1Department of Genetics, 2Department of Medicine, 3Department of Pediatrics, and 4Department of Microbiology and Immunology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA. 5Department of Pathology and Laboratory Medicine, Medical University of South Carolina, Charleston, South Carolina, USA. 6Center for Integrative Chemical Biology and Drug Discovery, University of North Carolina Eshelman School of Pharmacy, Chapel Hill, North Carolina, USA. 7Department of Medicine, The Dana Farber Cancer Institute, Harvard Medical School, Boston, Massachusetts, USA. 8Department of Toxicology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA.
Address correspondence to: Norman E. Sharpless, The Lineberger Comprehensive Cancer Center, University of North Carolina School Medicine, CB #7295, Chapel Hill, North Carolina 27599, USA. Phone: 919.966.1185; Fax: 919.966.8212; E-mail: nes@med.unc.edu.
Authorship note: Søren M. Johnson, Chad D. Torrice, and Jessica F. Bell contributed equally to this work.
Find articles by Johnson, S. in: JCI | PubMed | Google Scholar
1Department of Genetics, 2Department of Medicine, 3Department of Pediatrics, and 4Department of Microbiology and Immunology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA. 5Department of Pathology and Laboratory Medicine, Medical University of South Carolina, Charleston, South Carolina, USA. 6Center for Integrative Chemical Biology and Drug Discovery, University of North Carolina Eshelman School of Pharmacy, Chapel Hill, North Carolina, USA. 7Department of Medicine, The Dana Farber Cancer Institute, Harvard Medical School, Boston, Massachusetts, USA. 8Department of Toxicology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA.
Address correspondence to: Norman E. Sharpless, The Lineberger Comprehensive Cancer Center, University of North Carolina School Medicine, CB #7295, Chapel Hill, North Carolina 27599, USA. Phone: 919.966.1185; Fax: 919.966.8212; E-mail: nes@med.unc.edu.
Authorship note: Søren M. Johnson, Chad D. Torrice, and Jessica F. Bell contributed equally to this work.
Find articles by Torrice, C. in: JCI | PubMed | Google Scholar
1Department of Genetics, 2Department of Medicine, 3Department of Pediatrics, and 4Department of Microbiology and Immunology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA. 5Department of Pathology and Laboratory Medicine, Medical University of South Carolina, Charleston, South Carolina, USA. 6Center for Integrative Chemical Biology and Drug Discovery, University of North Carolina Eshelman School of Pharmacy, Chapel Hill, North Carolina, USA. 7Department of Medicine, The Dana Farber Cancer Institute, Harvard Medical School, Boston, Massachusetts, USA. 8Department of Toxicology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA.
Address correspondence to: Norman E. Sharpless, The Lineberger Comprehensive Cancer Center, University of North Carolina School Medicine, CB #7295, Chapel Hill, North Carolina 27599, USA. Phone: 919.966.1185; Fax: 919.966.8212; E-mail: nes@med.unc.edu.
Authorship note: Søren M. Johnson, Chad D. Torrice, and Jessica F. Bell contributed equally to this work.
Find articles by Bell, J. in: JCI | PubMed | Google Scholar
1Department of Genetics, 2Department of Medicine, 3Department of Pediatrics, and 4Department of Microbiology and Immunology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA. 5Department of Pathology and Laboratory Medicine, Medical University of South Carolina, Charleston, South Carolina, USA. 6Center for Integrative Chemical Biology and Drug Discovery, University of North Carolina Eshelman School of Pharmacy, Chapel Hill, North Carolina, USA. 7Department of Medicine, The Dana Farber Cancer Institute, Harvard Medical School, Boston, Massachusetts, USA. 8Department of Toxicology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA.
Address correspondence to: Norman E. Sharpless, The Lineberger Comprehensive Cancer Center, University of North Carolina School Medicine, CB #7295, Chapel Hill, North Carolina 27599, USA. Phone: 919.966.1185; Fax: 919.966.8212; E-mail: nes@med.unc.edu.
Authorship note: Søren M. Johnson, Chad D. Torrice, and Jessica F. Bell contributed equally to this work.
Find articles by Monahan, K. in: JCI | PubMed | Google Scholar
1Department of Genetics, 2Department of Medicine, 3Department of Pediatrics, and 4Department of Microbiology and Immunology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA. 5Department of Pathology and Laboratory Medicine, Medical University of South Carolina, Charleston, South Carolina, USA. 6Center for Integrative Chemical Biology and Drug Discovery, University of North Carolina Eshelman School of Pharmacy, Chapel Hill, North Carolina, USA. 7Department of Medicine, The Dana Farber Cancer Institute, Harvard Medical School, Boston, Massachusetts, USA. 8Department of Toxicology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA.
Address correspondence to: Norman E. Sharpless, The Lineberger Comprehensive Cancer Center, University of North Carolina School Medicine, CB #7295, Chapel Hill, North Carolina 27599, USA. Phone: 919.966.1185; Fax: 919.966.8212; E-mail: nes@med.unc.edu.
Authorship note: Søren M. Johnson, Chad D. Torrice, and Jessica F. Bell contributed equally to this work.
Find articles by Jiang, Q. in: JCI | PubMed | Google Scholar
1Department of Genetics, 2Department of Medicine, 3Department of Pediatrics, and 4Department of Microbiology and Immunology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA. 5Department of Pathology and Laboratory Medicine, Medical University of South Carolina, Charleston, South Carolina, USA. 6Center for Integrative Chemical Biology and Drug Discovery, University of North Carolina Eshelman School of Pharmacy, Chapel Hill, North Carolina, USA. 7Department of Medicine, The Dana Farber Cancer Institute, Harvard Medical School, Boston, Massachusetts, USA. 8Department of Toxicology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA.
Address correspondence to: Norman E. Sharpless, The Lineberger Comprehensive Cancer Center, University of North Carolina School Medicine, CB #7295, Chapel Hill, North Carolina 27599, USA. Phone: 919.966.1185; Fax: 919.966.8212; E-mail: nes@med.unc.edu.
Authorship note: Søren M. Johnson, Chad D. Torrice, and Jessica F. Bell contributed equally to this work.
Find articles by Wang, Y. in: JCI | PubMed | Google Scholar
1Department of Genetics, 2Department of Medicine, 3Department of Pediatrics, and 4Department of Microbiology and Immunology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA. 5Department of Pathology and Laboratory Medicine, Medical University of South Carolina, Charleston, South Carolina, USA. 6Center for Integrative Chemical Biology and Drug Discovery, University of North Carolina Eshelman School of Pharmacy, Chapel Hill, North Carolina, USA. 7Department of Medicine, The Dana Farber Cancer Institute, Harvard Medical School, Boston, Massachusetts, USA. 8Department of Toxicology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA.
Address correspondence to: Norman E. Sharpless, The Lineberger Comprehensive Cancer Center, University of North Carolina School Medicine, CB #7295, Chapel Hill, North Carolina 27599, USA. Phone: 919.966.1185; Fax: 919.966.8212; E-mail: nes@med.unc.edu.
Authorship note: Søren M. Johnson, Chad D. Torrice, and Jessica F. Bell contributed equally to this work.
Find articles by Ramsey, M. in: JCI | PubMed | Google Scholar
1Department of Genetics, 2Department of Medicine, 3Department of Pediatrics, and 4Department of Microbiology and Immunology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA. 5Department of Pathology and Laboratory Medicine, Medical University of South Carolina, Charleston, South Carolina, USA. 6Center for Integrative Chemical Biology and Drug Discovery, University of North Carolina Eshelman School of Pharmacy, Chapel Hill, North Carolina, USA. 7Department of Medicine, The Dana Farber Cancer Institute, Harvard Medical School, Boston, Massachusetts, USA. 8Department of Toxicology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA.
Address correspondence to: Norman E. Sharpless, The Lineberger Comprehensive Cancer Center, University of North Carolina School Medicine, CB #7295, Chapel Hill, North Carolina 27599, USA. Phone: 919.966.1185; Fax: 919.966.8212; E-mail: nes@med.unc.edu.
Authorship note: Søren M. Johnson, Chad D. Torrice, and Jessica F. Bell contributed equally to this work.
Find articles by Jin, J. in: JCI | PubMed | Google Scholar
1Department of Genetics, 2Department of Medicine, 3Department of Pediatrics, and 4Department of Microbiology and Immunology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA. 5Department of Pathology and Laboratory Medicine, Medical University of South Carolina, Charleston, South Carolina, USA. 6Center for Integrative Chemical Biology and Drug Discovery, University of North Carolina Eshelman School of Pharmacy, Chapel Hill, North Carolina, USA. 7Department of Medicine, The Dana Farber Cancer Institute, Harvard Medical School, Boston, Massachusetts, USA. 8Department of Toxicology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA.
Address correspondence to: Norman E. Sharpless, The Lineberger Comprehensive Cancer Center, University of North Carolina School Medicine, CB #7295, Chapel Hill, North Carolina 27599, USA. Phone: 919.966.1185; Fax: 919.966.8212; E-mail: nes@med.unc.edu.
Authorship note: Søren M. Johnson, Chad D. Torrice, and Jessica F. Bell contributed equally to this work.
Find articles by Wong, K. in: JCI | PubMed | Google Scholar
1Department of Genetics, 2Department of Medicine, 3Department of Pediatrics, and 4Department of Microbiology and Immunology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA. 5Department of Pathology and Laboratory Medicine, Medical University of South Carolina, Charleston, South Carolina, USA. 6Center for Integrative Chemical Biology and Drug Discovery, University of North Carolina Eshelman School of Pharmacy, Chapel Hill, North Carolina, USA. 7Department of Medicine, The Dana Farber Cancer Institute, Harvard Medical School, Boston, Massachusetts, USA. 8Department of Toxicology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA.
Address correspondence to: Norman E. Sharpless, The Lineberger Comprehensive Cancer Center, University of North Carolina School Medicine, CB #7295, Chapel Hill, North Carolina 27599, USA. Phone: 919.966.1185; Fax: 919.966.8212; E-mail: nes@med.unc.edu.
Authorship note: Søren M. Johnson, Chad D. Torrice, and Jessica F. Bell contributed equally to this work.
Find articles by Su, L. in: JCI | PubMed | Google Scholar
1Department of Genetics, 2Department of Medicine, 3Department of Pediatrics, and 4Department of Microbiology and Immunology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA. 5Department of Pathology and Laboratory Medicine, Medical University of South Carolina, Charleston, South Carolina, USA. 6Center for Integrative Chemical Biology and Drug Discovery, University of North Carolina Eshelman School of Pharmacy, Chapel Hill, North Carolina, USA. 7Department of Medicine, The Dana Farber Cancer Institute, Harvard Medical School, Boston, Massachusetts, USA. 8Department of Toxicology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA.
Address correspondence to: Norman E. Sharpless, The Lineberger Comprehensive Cancer Center, University of North Carolina School Medicine, CB #7295, Chapel Hill, North Carolina 27599, USA. Phone: 919.966.1185; Fax: 919.966.8212; E-mail: nes@med.unc.edu.
Authorship note: Søren M. Johnson, Chad D. Torrice, and Jessica F. Bell contributed equally to this work.
Find articles by Zhou, D. in: JCI | PubMed | Google Scholar
1Department of Genetics, 2Department of Medicine, 3Department of Pediatrics, and 4Department of Microbiology and Immunology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA. 5Department of Pathology and Laboratory Medicine, Medical University of South Carolina, Charleston, South Carolina, USA. 6Center for Integrative Chemical Biology and Drug Discovery, University of North Carolina Eshelman School of Pharmacy, Chapel Hill, North Carolina, USA. 7Department of Medicine, The Dana Farber Cancer Institute, Harvard Medical School, Boston, Massachusetts, USA. 8Department of Toxicology, The Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA.
Address correspondence to: Norman E. Sharpless, The Lineberger Comprehensive Cancer Center, University of North Carolina School Medicine, CB #7295, Chapel Hill, North Carolina 27599, USA. Phone: 919.966.1185; Fax: 919.966.8212; E-mail: nes@med.unc.edu.
Authorship note: Søren M. Johnson, Chad D. Torrice, and Jessica F. Bell contributed equally to this work.
Find articles by Sharpless, N. in: JCI | PubMed | Google Scholar
Authorship note: Søren M. Johnson, Chad D. Torrice, and Jessica F. Bell contributed equally to this work.
Published June 23, 2010 - More info
J Clin Invest. 2010;120(7):2528–2536. https://doi.org/10.1172/JCI41402.
© 2010 The American Society for Clinical Investigation
Received: October 12, 2009; Accepted: April 28, 2010
Related article:
Abstract
The efficacy of cancer treatment by radiation and chemotherapeutic drugs is often limited by severe side effects that primarily affect the hematopoietic system and the epithelium of the gastrointestinal tract. Progress in understanding differences in the mechanisms involved in the responses of normal and tumor cells to genotoxic stress has led to the development of new rational approaches to selective protection of normal cells, such as suppression of apoptosis by pharmacological inhibition of p53 or activation of NF-κB. Another promising approach presented in this issue by Johnson et al. is based on the idea of using pharmacological inhibitors of cyclin-dependent kinases (CDKs) to convert normal cells into a radioresistant state by inducing reversible cell cycle arrest at the G1/S transition. The evidence indicates that this approach is likely to be specific for protection of normal cells and may, therefore, have clinical potential as an adjuvant in anticancer therapies.
Authors
Andrei V. Gudkov, Elena A. Komarova
-
Abstract
Total body irradiation (TBI) can induce lethal myelosuppression, due to the sensitivity of proliferating hematopoietic stem/progenitor cells (HSPCs) to ionizing radiation (IR). No effective therapy exists to mitigate the hematologic toxicities of TBI. Here, using selective and structurally distinct small molecule inhibitors of cyclin-dependent kinase 4 (CDK4) and CDK6, we have demonstrated that selective cellular quiescence increases radioresistance of human cell lines in vitro and mice in vivo. Cell lines dependent on CDK4/6 were resistant to IR and other DNA-damaging agents when treated with CDK4/6 inhibitors. In contrast, CDK4/6 inhibitors did not protect cell lines that proliferated independently of CDK4/6 activity. Treatment of wild-type mice with CDK4/6 inhibitors induced reversible pharmacological quiescence (PQ) of early HSPCs but not most other cycling cells in the bone marrow or other tissues. Selective PQ of HSPCs decreased the hematopoietic toxicity of TBI, even when the CDK4/6 inhibitor was administered several hours after TBI. Moreover, PQ at the time of administration of therapeutic IR to mice harboring autochthonous cancers reduced treatment toxicity without compromising the therapeutic tumor response. These results demonstrate an effective method to mitigate the hematopoietic toxicity of IR in mammals, which may be potentially useful after radiological disaster or as an adjuvant to anticancer therapy.
-
Introduction
The cytotoxicity of DNA-damaging agents such as ionizing radiation (IR) is cell cycle dependent. In particular, early G1 and late S phases are relatively radioresistant, whereas the G1/S transition and G2/M phases are relatively radiosensitive (1, 2). Traversing from G1 to S phase, while harboring DNA damage, is particularly toxic (3), and an extended period of G1 after exposure to genotoxins enhances resistance (4–6), possibly by allowing for greater DNA repair prior to G1/S traversal. We therefore reasoned that modulating the cell cycle by lengthening G1 may mitigate the toxicity of DNA-damaging agents such as IR.
The transition from G1 to S phase is regulated by at least 3 cyclin-dependent kinases (CDK2, CDK4, and CDK6) and their catalytic partner cyclins (cyclin A, cyclin D, and cyclin E), functioning in concert to phosphorylate the Rb family proteins (reviewed in ref. 7). Combinations of CDK-cyclin gene knockouts in mice have revealed considerable redundancy and/or developmental compensation among these proteins (reviewed in ref. 8). For example, murine embryos lacking both CDK4 and CDK6 or all the D-type cyclins demonstrate normal proliferation in many tissues but succumb late in development, due to a lack of fetal hematopoietic stem/progenitor cells (HSPCs) (9, 10). Likewise, in adult mice, pharmacologic inhibition of CDK4/6 activity is well tolerated, only inhibiting proliferation in a few CDK4/6-dependent cell types (11). Importantly, CDK2 and CDK1 phosphorylate several non-Rb substrates that regulate additional processes, such as transcription and DNA metabolism (reviewed in ref. 12), whereas CDK4/6 are highly specific to Rb family proteins. Based on these observations, we hypothesized that we could improve resistance to DNA-damaging agents by inducing a G1 arrest using selective pharmacologic CDK4/6 inhibitors.
-
Results
To test this hypothesis in vitro, we first screened putative CDK4/6 inhibitors for their cell cycle effects in several human cell lines. CDK4/6-dependent cell lines, including telomerized human diploid fibroblasts (tHDFs) and a human melanoma cell line (WM2664), demonstrated strong, reversible G1 arrest (pharmacological quiescence [PQ]) after exposure to the potent and selective CDK4/6 inhibitors, PD0332991 (13, 14) and 2BrIC (15), whereas the less selective CDK inhibitors that additionally target CDK1/2 (roscovitine [ref. 16], R547 [ref. 17], and flavopiridol [ref. 18]) variably produced a G2/M block, intra-S arrest, or cell death in these cell types (data not shown and Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI41402DS1). Effects of the nonselective CDK inhibitors were similar on an Rb-null melanoma line, A2058, whereas the selective CDK4/6 inhibitors had no effect on A2058 cell cycle or survival, even at doses well above the IC50 in CDK4/6-dependent cell lines (Supplemental Figure 1). The proliferation of 7 Rb-deficient human small cell lung cancer lines was also resistant to CDK4/6 inhibitors (data not shown), again reinforcing the notion that Rb-null cells are generally unaffected by CDK4/6 inhibition. These data show that structurally distinct, potent, and selective CDK4/6 inhibitors effect a “clean” G1 arrest — without cytotoxicity, intra-S or G2 arrest — in susceptible cell lines, whereas the cell cycle effects of more global CDK inhibitors are less predictable and associated with cytotoxicity.
Having identified 2 highly selective CDK4/6 inhibitors that elicit a clean G1 arrest in dependent cells, we next sought to determine whether PQ was protective from cell cycle–dependent genotoxic events in CDK4/6-dependent cell lines. IR exposure (Figure 1) and doxorubicin exposure (Figure 2) both caused extensive DNA damage (phospho–γ-H2AX foci and DNA strand breaks), elicited a robust DNA damage response (phospho-p53 and p21CIP expression), and caused cell death (WST-1 assay) in all 3 cell lines. Treatment with PD0332991 or 2BrIC prior to genotoxic stress attenuated γ-H2AX formation (Figure 1A, Figure 2B, and Supplemental Figure 2, A and B), DNA strand breaks (Figure 1C and Supplemental Figure 2C), and DNA damage response (Figure 1B, Figure 2C, and Supplemental Figure 2D) and promoted cellular survival in the CDK4/6-dependent cell lines WM2664 and tHDF (Figure 2D and Supplemental Figure 2E and Supplemental Figure 3A). Similar protection was observed after exposure to another DNA-damaging agent, etoposide (data not shown). In contrast, no protective effects of PD0332991 or 2BrIC treatment were noted in Rb-deficient A2058 cells (Figure 2D and Supplemental Figure 2E and Supplemental Figure 3A), and the less selective CDK inhibitors (roscovitine, flavopiridol, and R547) failed to enhance cell survival in any cell line (Supplemental Figure 3, B–D). The failure of the less selective inhibitors to afford protective PQ suggests that arrest in a phase of the cell cycle other than G1 (e.g., G2/M) may not protect from genotoxic exposure. Alternatively, the less selective CDK inhibitors may prevent phosphorylation of non-Rb family substrates by CDK2 (e.g., BRCA1 [ref. 19] or CtIP [ref. 20]) and thereby untowardly augment the toxicity of DNA-damaging agents. Together, these data show that PQ, effected by selective CDK4/6 inhibitors but not more global CDK inhibitors, provides in vitro resistance to DNA-damaging agents in cell types that require CDK4/6 kinase activity for G1 to S traversal.
 Figure 1
Figure 1PQ enhances radioresistance in vitro. (A) Images of phospho–γ-H2AX foci (green) and phalloidin staining (red) of tHDFs, with or without 6 Gy IR and 24 hours of 100 nM PD0332991 exposure. Original magnification, ×40. Scale bar: 50 μm. (B) Western blots showing phospho-p53 (p-p53; arrow) induction in tHDF lysates after 6 Gy IR, with and without PD0332991 treatment. (C) Comet tail area, after 24 hours of exposure to 100 nM PD0332991, prior to the indicated IR dose. Forty cells in each condition were imaged at an original magnification of ×20. *P < 0.01, **P < 0.0001, for pair-wise comparisons.
 Figure 2
Figure 2PQ enhances radioresistance in vitro. (A) An in vitro schedule of CDK inhibitor (CDKi) and doxorubicin (Dox) exposure using tHDFs. Assays were performed where indicated on the timeline with arrows. WB, Western blot. (B) Flow cytometry analysis of phospho–γ-H2AX formation after PD0332991 and doxorubicin treatment of tHDFs. (C) Western blot analysis of DNA damage response markers in tHDFs, at indicated times after PD0332991 treatment, with or without doxorubicin treatment. The black bar indicates lysates harvested immediately after IR exposure (t = 0 hours), and the gray bar indicates lysates harvested 12 hours after IR exposure (t = 12 hours). (D) WST assay for cell proliferation at 7 days after doxorubicin exposure. All absorbances are normalized to DMSO. #P < 0.01 vs. DMSO, ##P < 0.001 vs. DMSO, *P < 0.01 vs. doxorubicin, **P < 0.001 vs. doxorubicin, ***P < 0.0001, for the indicated pair-wise comparisons. “Wash” indicates time in media without CDK inhibitor or doxorubicin, as shown in A.
To further test whether the protective effects of selective CDK4/6 inhibitors required the induction of G1 arrest, we performed cell synchronization experiments. CDK4/6-dependent cells were released from CDK4/6 inhibition and allowed to progress to G2/M prior to doxorubicin exposure (Figure 2A, schematic). Under such conditions, tHDFs and WM2664 cells demonstrated a marked increase in cellular toxicity as measured by increased γ-H2AX foci, increased DNA damage response, and decreased cellular survival (Figure 2, B–D). Therefore, cells benefit from CDK4/6 inhibition only when it results in a G1 arrest, and cells in G2 have enhanced sensitivity to DNA damage. This conclusion is supported by the observation that other means of promoting a G1 arrest (e.g., staurosporine treatment in certain cell types; ref. 21) can also enhance protection from genotoxic agents.
These in vitro data suggest that CDK4/6-dependent tissues in vivo might also be protected from DNA-damaging agents by CDK4/6 inhibitors. Motivated by prior work showing that fetal hematopoietic progenitors require CDK4/6–cyclin D for proliferation (9, 10) as well as the clinical need to protect the hematopoietic system from DNA-damaging agents, we sought to determine whether hematopoietic progenitors could be rendered pharmacologically quiescent in vivo. Proliferation of murine HSCs (Lin–Kit+Sca1+CD48–CD150+), as measured by Ki67 expression and incorporation of BrdU over 24 hours (Figure 3), was comparable to prior estimates (22, 23). When PD0332991, which is orally bioavailable, was administered for 48 hours prior to bone marrow harvest, the frequency of HSC proliferation decreased significantly (Figure 3, B and C), with the greatest effect observed in Ki67 expression. An even more pronounced inhibition of proliferation was noted in the more rapidly proliferating multipotent progenitor (MPP) cell compartment (Lin–Kit+Sca1+CD48–CD150–). Oligopotent progenitors (Lin–Kit+Sca1–) demonstrated a modest inhibition of proliferation, with the strongest effects seen in common myeloid progenitors (CMPs) and common lymphocyte progenitors (CLPs), compared with weaker effects in the more differentiated granulocyte-monocyte progenitors (GMPs) and megakaryocyte-erythroid progenitors (MEPs; Figure 3, B and C). In contrast to these effects in early HSPCs, no change in proliferation was noted in the more fully differentiated Lin–Kit–Sca1– and Lin+ cells, though these fractions are heterogeneous and effects on subpopulations may be obscured.
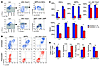 Figure 3
Figure 3CDK4/6 inhibition decreases HSPC proliferation. (A) Flow cytometry gating scheme for HSCs and MPPs (top) and myeloid progenitors (bottom) using cell surface antigens. Gray regions represent cell populations used for further analysis as indicated. FSc, forward scatter. (B) Representative contour plots of proliferation in indicated HSPC populations, as measured by BrdU incorporation and Ki67 expression, after 48 hours of no treatment (n = 6) or PD0332991 treatment (n = 6) in the presence of BrdU for 24 hours. Contours represent 5% density. (C) Quantification of BrdU and Ki67 data in all HSPC subpopulations. (D) Relative frequency of Lin–, HSC, MPP, or Lin–cKit+ Sca1– populations after 48 hours of PD0332991 treatment and 24 hours of BrdU exposure. *P < 0.05, **P < 0.01, ***P < 0.001.
These effects on immunophenotypic HSPC frequency were validated by other methods. Transient (48-hour) treatment with PD0332991 did not decrease total marrow cellularity (Supplemental Figure 4A) but did decrease the absolute number of lineage-negative cells (Figure 3D) without altering HSPC apoptosis or viability (Supplemental Figure 4, B and C). The frequency of the more abundant oligopotent progenitors declined (Lin–cKit+Sca1–; Figure 3D and Supplemental Figure 4D), with an associated relative increase in HSC and MPP frequencies. Cobblestone area–forming cell (CAFC) assays confirmed that transient CDK4/6 inhibition did not decrease the in vivo HSPC number (Supplemental Figure 4E). In combination, these data suggest a gradient of dependence on CDK4/6 activity for proliferation during myeloid/erythroid differentiation: the least differentiated cells (HSCs, MPPs, and CMPs) appeared to be the most dependent, more differentiated elements (GMPs and MEPs) were less dependent, and even more differentiated myeloid and erythroid cells proliferated independently of CDK4/6 activity.
In accordance with this model, serial daily treatment with PD0332991 for 12 days caused a modest decrease in the erythroid, platelet, and myeloid (monocyte and granulocyte) lineages, which only became apparent after 8 days of treatment, and which began to improve within 4 days, upon cessation of PD0332991 (Supplemental Figure 5). These observations are consistent with the kinetics and degree of myelosuppression seen when tumor-bearing mice (11) and human patients with malignancies (24, 25) are serially treated with PD0332991. These data confirm that the short-term production of differentiated peripheral blood effector cells is relatively resistant to CDK4/6 inhibition and that the myelosuppressive effects of CDK4/6 inhibition are rapidly reversible in vivo.
Given that PQ affords cellular protection in vitro (Figures 1 and 2) and effects a specific G1 arrest of HSPCs in vivo (Figure 3), we next sought to test whether CDK4/6 inhibition would render wild-type mice resistant to the hematological toxicity of total body irradiation (TBI), administered at doses around the LD90 (7.5 Gy). Adult female C57BL/6 mice were exposed to 6.5, 7.5, or 8.5 Gy, with or without PD0332991 treatment (administered as shown in Figure 4A), and followed for 40 days after TBI. Marked protection from the hematological toxicity of peri-lethal doses of TBI was observed. Nearly all untreated mice succumbed to death from hematological toxicity when exposed to 7.5 Gy, whereas all treated mice survived (Figure 4B). At 8.5 Gy TBI, PD0332991 treatment significantly increased the 30-day survival (13% treated vs. 0% untreated mice) and prolonged the median survival (19 days for treated vs. 13 days for untreated; Supplemental Figure 6A). All mice survived after a dose of 6.5 Gy, regardless of PD0332991 treatment (Supplemental Figure 6B). These data show that PQ resulting from transient CDK4/6 inhibition around the time of TBI enhances radioresistance in vivo.
 Figure 4
Figure 4PQ at the time of TBI increases survival. (A) The PD0332991 treatment schedule in initial radioprotection experiments. The arrows indicate doses of PD0332991 before or after TBI (shown as lightning bolt). (B) Kaplan-Meier analysis of survival after 7.5 Gy of TBI, with or without PD0332991 treatment. (C) Survival after 7.5 Gy TBI, with the indicated dosing schedules of PD0332991. (D) Aggregate results of animals gavaged 4 hours prior to TBI, representing results from 3 murine strains with animals of both sexes. Results of strains considered independently were comparable in all groups (see Figure 6C and Supplemental Figure 6C). All survival curve P values were calculated using the log-rank test as a pair-wise comparison with the untreated group. (E) Caspase activation in BM-MNCs after exposure to 7.5 Gy TBI (x-rays; see Methods). (F) Complete blood counts with differential at 21 days after 7.5 Gy TBI, with and without PD0332991 treatment. Treated animals received PD0332991 by oral gavage at 28 and 4 hours prior to (–28 and –4, respectively) and 20 hours after (+20) IR dose on day 0. Myeloid cells include granulocytes and monocytes, and # indicates that the maximum value of the cohort is shown in lieu of error bars where cells numbers were too small to reliably quantify. *P < 0.05, **P < 0.01, ***P < 0.001.
Because 3 doses of PD0332991 at different times relative to TBI provided radioprotection, we next sought to determine which of these doses was most important for radioprotection. We observed that most of the benefit of the treatment schedule was associated with CDK4/6 inhibitor treatment immediately prior to or simultaneous with TBI at the LD90. Mice treated with a single dose 4 hours before or a single dose at TBI (time 0) demonstrated survival similar to that of animals treated on the multidose schedule (doses at 28 and 4 hours before TBI and 20 hours after TBI) (Figure 4, B and C). The benefit of treatment at 4 hours prior to TBI was confirmed in a larger cohort of mice (n = 62), including matched animals from 3 different inbred strains (C57BL/6, C3H, and FVB/n) and both male and female mice (Figure 4D). Surprisingly, even a single dose 20 hours after TBI significantly enhanced survival after TBI (Figure 4C). As therapeutic serum levels are not achieved until more than 30 minutes after gavage and then persist for 10–20 hours (data not shown), these observations suggest that a period of PQ lasting for several hours (>20 hours) after the induction of DNA damage is beneficial. Although there are a few known compounds that protect from radiotoxicity when administered prior to IR (i.e., “radioprotectants”) (26, 27), we are unaware of any hematological “radiomitigants,” (i.e., compounds that reduce myelotoxicity when administered many hours after an exposure to TBI).
TBI at doses used in this study causes morbid pancytopenia, with count nadirs occurring 14–21 days after TBI (28). To confirm that the improved survival afforded by PQ was due to protection of hematopoietic lineages, we examined the bone marrow and peripheral blood of animals after IR exposure. We observed a marked reduction of caspase activity in bone marrow mononuclear cells (BM-MNCs) harvested 16 hours after TBI from mice that had been pretreated with PD0332991 prior to TBI (Figure 4E). In accordance with this protection observed in the bone marrow after TBI, we also observed that PD0332991 treatment significantly ameliorated pancytopenia in lethally irradiated mice (Figure 4F). A similar improvement of cell count nadirs and more rapid cell count recovery was observed after a sublethal dose of TBI (Supplemental Figure 7). Importantly, PQ therapy had a beneficial effect on the recovery of all peripheral blood lineages: platelets, erythrocytes, myeloid cells (granulocytes and monocytes), and peripheral lymphocytes. The improvement in quadrilineage hematopoiesis after TBI is consistent with the notion that CDK4/6 inhibition exerts maximal radioprotection in the early HSPCs rendered quiescent by CDK4/6 inhibitor treatment.
In order to bolster these pharmacologic data, we next turned to a genetic model. Prior work in mice has shown that p21CIP, a potent CDK inhibitor and critical mediator of the p53-mediated DNA damage response (29, 30), plays an important role in regulating HSPC quiescence after DNA damage (31). To test the importance of p21CIP in hematologic radiomitigation, we exposed inbred p21CIP-null, heterozygous, and wild-type littermates to lethal TBI, both without (Figure 5A) and with (Figure 5B) PD0332991. For these experiments, an x-ray IR source was used at a dose greater than the LD90. Confirming the critical role of p21CIP in mediating cellular survival after IR, survival after TBI strongly correlated with p21CIP gene dosage: p21CIP-null animals were most sensitive, while wild-type animals were most resistant, and heterozygotes were intermediate (Figure 5A). Administration of PD0332991 4 hours prior to TBI improved survival in all 3 genotypes, but PD0332991 treatment did not fully rescue the radiosensitivity of p21CIP-null mice (Figure 5B). These observations are in accordance with PQ data (Figure 4C), suggesting that the induction of a cell cycle pause well after the onset of DNA damage improves HSPC survival. Moreover, the beneficial effects of PQ after TBI do not strictly require p21CIP, although expression of this CDK inhibitor appeared to enhance the effects of PQ.
 Figure 5
Figure 5Cell cycle arrest after lethal TBI improves survival and requires both p21CIP and CDK4/6 inhibition for optimal benefit. Kaplan-Meier survival curves for wild-type, heterozygous, and p21CIP-null mice, exposed to lethal TBI 4 hours after oral gavage with (A) vehicle or (B) PD0332991. Mice were irradiated using an x-ray source (see Methods). The log-rank test for trend is shown.
Prior work has suggested that some radioprotectant strategies are associated with an increased late risk of leukemia (32), presumably resulting from an augmented survival of damaged cells after IR. To assess the risk of late toxicity, we serially examined mice after TBI in the presence or absence of PQ. When followed 210–274 days after TBI, no deaths were seen in any animals after 6.5 Gy TBI, regardless of PD0332991 treatment. Only 2 out of 18 mice (1 C3H and 1 C57BL/6) survived 7.5 Gy TBI in the absence of PQ, and these animals showed no evidence of disease 143–252 days after TBI. Of 29 mice surviving the acute toxicity of 7.5 Gy TBI in the setting of PQ, there was 1 death of unknown cause at day 99 after TBI, and the remaining mice were disease free 101–251 days after TBI. Blood counts of long-term–surviving animals were comparable among unirradiated and irradiated mice, with or without PD0332991 treatment, at the time of TBI (Supplemental Figure 8). No evidence of myeloproliferative disorder or myelodysplasia was seen in animals of these long-term–surviving cohorts. These data indicate that PQ does not exacerbate the late hematological toxicity associated with sublethal TBI and affords good long-term hematological radioprotection, even after a lethal dose of TBI.
Although these results demonstrate the ability of PQ to reduce the toxicity of DNA-damaging agents, exposure to IR and other DNA-damaging agents more commonly occurs therapeutically in the treatment of cancer. To determine whether PQ could protect the bone marrow of a tumor-bearing host without also protecting its cancer, we examined the effects of PQ on tumor response in a well-defined, genetically engineered murine model of melanoma (33). In this model, male TyrRas Ink4a/Arf–/– mice develop autochthonous melanomas, driven by melanocyte-specific expression of mutant H-Ras in the setting of combined p16INK4a and Arf inactivation (Figure 6A). Even though development of these tumors is facilitated by loss of p16INK4a (34), a potent CDK4/6 inhibitor, their growth was not inhibited by serial CDK4/6 inhibitor treatment (Figure 6B), suggesting established tumors in this model do not require CDK4/6 activity for continued proliferation. Moreover, these tumors are sensitive to IR, with 7.5 Gy TBI arresting tumor growth for approximately 20 days after IR, whereas all unirradiated tumors demonstrated overt progression within 10 days (Figure 6B). However, when tumor-bearing mice were treated with a single dose of PD0332991 4 hours prior to TBI, an impressive rescue of radiation-induced morbidity, in terms of weight loss (Supplemental Figure 9B) and mortality (Figure 6C), was seen without a change in tumor response (Figure 6, B and C). These data reveal that PQ can reduce treatment-associated myelosuppression after exposure to DNA-damaging agents, without concomitantly protecting an established, CDK4/6-independent cancer.
 Figure 6
Figure 6CDK4/6 inhibition improves survival without compromising tumor sensitivity to radiation therapy. (A) Representative images of the progression of an autochthonous TyrRas Ink4a/Arf–/– melanoma, despite daily oral therapy with PD0332991. (B) Tumor growth with or without continuous daily PD0332991 treatment compared with tumor growth with or without PD0332991 administered as a single dose 4 hours prior to 7.5 Gy TBI. “NS” is nonsignificant for the comparison between groups receiving 7.5 Gy TBI. (C) Kaplan-Meier survival curves showing overall mortality and mortality subdivided by cause. P values were calculated using the log-rank test.
-
Discussion
The cell cycle phase has long been linked to cellular radiosensitivity (1, 2), and attempts to modify radiosensitivity through manipulation of the cell cycle were originally described several decades ago (35, 36). The approach described in the present work adds specificity for HSPCs through selective pharmacologic inhibition of CDK4/6. Our data demonstrate that such inhibition induced a G1 arrest in CDK4/6-dependent cells (Supplemental Figure 1), thereby protecting such cells from genotoxins. In contrast, cells that did not require CDK4/6 activity for proliferation were not arrested by CDK4/6 inhibitors and were not protected by such agents (Figures 1 and 2). Likewise, less selective CDK inhibitors that target CDK1/2, among other kinases, did not induce a clean G1 arrest in vitro and did not afford protection from DNA-damaging agents. If anything, in our studies, nonselective CDK inhibitors appear to augment the toxicity of DNA-damaging agents, in accordance with prior work from several other groups (20, 37, 38).
Although the precise mechanism(s) by which an augmented G1 arrest protects cells from IR cytotoxicity is not well understood, our data support a few possible explanations. We show that cells arrested in G1 demonstrate an increased resistance to the DNA-damaging effects of IR (Figure 1). Other groups have reported an intrinsic resistance to DNA-damaging agents in G1 as well as an augmented rate of DNA repair in this phase of the cell cycle (39, 40). A reduced susceptibility to IR-induced DNA damage in G1 could partially explain the radioprotection we observed; however, such an explanation cannot account for the radiomitigation effects of PQ in vivo, where G1 arrest after IR exposure is protective.
Prevention of G1 to S traversal is an alternative mechanism that can explain the radiomitigation effects of CDK4/6 inhibitors. Attempted G1 to S traversal, in the setting of unrepaired DNA damage, is a particularly toxic event (3, 41), likely due to activation of early S phase transcription factors, such as the E2Fs, which regulate many effectors of apoptosis (42, 43). This mechanism is consistent with increased radiosensitivity noted in late G1 (1, 2), more recently described as damage occurring after the G1 restriction point (44, 45). Additionally, it may be that certain types of IR-induced lesions (single-strand breaks [SSBs] or more rarely interstrand crosslinks [ISCLs]) are relatively nontoxic in G1 (46) but become more toxic in S phase, as these lesions are converted to toxic DSBs (e.g., through stalled replication forks). Indeed, although DSBs are highly correlated with cytotoxicity, approximately 90% of DNA lesions after IR are SSBs (47), while ISCLs may also be induced at lower frequency by IR (48). Therefore, inhibiting G1 to S traversal may be beneficial, by preventing apoptosis associated with E2F induction or by allowing time for repair of lesions that are well tolerated during G1, but which may be converted to more toxic DSBs during S phase.
We believe prevention of G1 to S traversal explains how selective CDK4/6 inhibitors improve survival when administered concurrent with or up to 20 hours after exposure to lethal IR (Figure 4C). This mechanism is additionally supported by the requirement for induction of p21CIP for optimal survival after IR (Figure 5). Expression of p21CIP peaks 24–48 hours after IR exposure (49), and mice lacking p21CIP are sensitive to the hematologic toxicities of IR (Figure 5). Therefore, a cell cycle pause induced by p21CIP hours after IR ameliorates the hematopoietic toxicity of IR. Additional support for this view is provided by the recent finding that FBXO31 expression is required for maximal radioprotection after IR (50). Like p21CIP, FBXO31 expression is induced hours after IR in an ATM-dependent manner, and its expression induces degradation of cyclin D1, thereby reducing CDK4/6 activity and inducing a G1 arrest in some cell types. Therefore, a maximal G1 arrest effected by disparate mechanisms after IR appears beneficial for survival. Additionally, the G1 arrest induced in HSPCs by antiproliferative mechanisms, like p21CIP, after IR does not appear complete, in that it can be further enhanced by PQ (Figure 5).
Current clinical interventions to mitigate radiation toxicity have relied on a combination of supportive care, growth factors, cytokines, and specific chelating agents — none of which are effective when administered several hours after radiation exposure (reviewed in ref. 27). While growth factor support with agents such as G/GM-CSF or erythropoietin has been shown to attenuate the toxic effects of DNA-damaging agents (32, 51), the small molecule PQ approach appears superior in magnitude of effect, has a longer effective duration after exposure, and protects without the significant expense and toxicities of these biologics. Small molecule CDK4/6 inhibitors are orally bioavailable and can be readily stockpiled, both highly desirable features for agents to be used in the setting of a radiologic disaster. Moreover, PQ ameliorates DNA damage–induced thrombocytopenia (Figure 4F and Supplemental Figure 7), a significant, unmet need in clinical oncology and radiation mitigation. While late hematological toxicity has been associated with growth factor support after exposure to DNA-damaging agents in both humans (52, 53) and mice (32), PQ does not appear to augment late hematological toxicity after TBI (Supplemental Figure 8). Furthermore, growth factor support and PQ appear to enhance count recovery through different mechanisms: the former by inhibiting apoptosis, increasing HSPC proliferation, and modulating lineage choice; the latter by promoting DNA repair prior to a resumption of progenitor proliferation. Therefore, we think it likely that the combined use of PQ and growth factor support may prove particularly beneficial in ameliorating myelosuppression resulting from the exposure to DNA-damaging agents.
In addition to providing radiomitigation, PQ could also be used as an adjuvant to clinical anticancer therapy. Although cell cycle alteration has successfully improved survival from lethal IR in the past (35, 36), prior strategies possessed limited clinical utility due to the lack of selective alteration of the cell cycle in the bone marrow without affecting tumors. The PQ approach capitalizes upon the relatively infrequent requirement for CDK4/6 activity for tumor maintenance in order to selectively radioprotect hematopoietic progenitors (Figure 4, E and F), thereby improving survival after IR without compromising tumor cell kill (Figure 6, B and C). Preventing myelosuppression after cytotoxic cancer therapies would allow increased dose intensity and density, allowing increased tumor cell kill. However, our data do suggest that PQ would compromise tumor cell kill in cancers that are sensitive to CDK4/6 inhibition, although we believe this concern would not apply to most human malignancies. For example, CDK4/6 inhibitors failed to induce G1 arrest in Rb-deficient small cell lung cancer cell lines, and therefore PQ would be unlikely to compromise chemotherapeutic efficacy in the approximately 10% of human cancers that are Rb deficient. A similar logic applies to cancers with other genetic lesions that render them insensitive to CDK4/6 inhibition (e.g., Myc activation). Importantly, even some tumors (e.g., TyrRas Ink4a/Arf–/– melanoma) whose progression is facilitated by inactivation of p16INK4a, an endogenous CDK4/6 inhibitor, were not protected by CDK4/6 inhibitors at the time of IR exposure (Figure 6B). Therefore, it is likely that PQ could attenuate the hematological toxicity of anticancer therapies in a substantial fraction of human cancer patients without a concomitant diminution in anticancer efficacy.
CDK4/6 inhibitors are in an advanced stage of clinical development as antineoplastics. PD0332991 has been administered to humans (24, 25), and 5 current phase I/II trials with this compound are planned or underway testing (listed at http://clinicaltrials.gov/ as NCT00555906, NCT00420056, NCT00721409, NCT00141297, and NCT01037790). Several other series of selective CDK4/6 inhibitors have been reported (54), with at least 2 additional candidates slated to soon enter human phase I trials. Therefore, we believe the ready availability of these relatively nontoxic, small molecule inhibitors with favorable pharmacology in humans will allow for human testing of the beneficial effects of PQ after TBI and DNA-damaging agents in short order.
-
Methods
Animals. All animal experiments were performed with approval of the University of North Carolina Institutional Animal Care and Use Committee. For studies of wild-type mice, young adult (8–12 weeks of age) C57BL/6 (The Jackson Laboratory) or C3H (Harlan Sprague-Dawley) animals were used. TyrRas Ink4a/Arf–/– mice were FVB/n background, and p21CIP–/– mice were obtained from Hanno Hock (Massachusetts General Hospital, Boston, Massachusetts, USA) and then fully backcrossed (n = 10 generations) to C57BL/6 for study. Peripheral blood was collected by tail vein nick for complete blood counts (HemaTrue, Heska). Experiments on tumor-bearing TyrRAS+ Ink4a/Arf–/– mice (33) were performed in males fully backcrossed (n > 10) to the FVB/n background. Mice were treated as previously described (11) with PD0332991 (Pfizer Inc.), given by oral gavage at a dose of 150 mg/kg BW. TyrRAS+ Ink4a/Arf–/– mice were serially observed for tumor development. When tumors were noted to be approximately 0.2 cm2 in size, animals were treated as described and tumor response was assessed by daily caliper measurements. Data in Figure 6B are normalized to tumor size at the time of therapy initiation, with volumes calculated using the formula (width × length)2/2. Tumor-bearing mice were euthanized at the indicated times for morbidity, tumor ulceration, or tumor size of more than 1.5 cm in diameter.
IR sources. Animals were irradiated using a 137Cs AECL GammaCell 40 Irradiator (Atomic Energy of Canada) or a XRAD320 (Precision XRay Inc.) biological irradiator. Experiments were carried out using the 137Cs source, unless otherwise noted. Our empirically determined LD90 in this system was 7.5 Gy, consistent with prior studies (28, 32, 51, 55).
Cell lines, γ-H2AX by flow cytometry, WST-1, and Western blots. tHDFs (from Gordon Peters, London Research Institute, London, United Kingdom) were cultured in DMEM plus 10% FBS with any additional compounds. The same conditions were used for A2058 and WM2664, human melanoma cell lines with known Rb-pathway mutations: A2058 is Rb-null, whereas WM2664 lacks Ink4a/Arf (56). For γ-H2AX assay, cells were fixed, permeabilized, and stained with anti–γ-H2AX as per the γ-H2AX Flow Kit (Millipore). Cell viability was assessed using a colorimetric assay for cleavage of water soluble tetrazolium salt (WST-1 assay; TaKaRa Bio USA), performed by seeding 1 × 103 cells per well in a 96-well tissue culture plate in 100 μl of growth medium and treating as indicated with CDK4/6 inhibitor (100 nM PD0332991 or 2 μM 2BrIC) and radiomimetic (1 μM doxorubicin). Following treatment, cells were allowed to recover for 7 days in normal growth medium, and then the cell number was quantified using the WST-1 assay. For cell number and viability, as measured by CellTiter-Glo Luminescent Cell Viability Assay (Promega), cells were seeded overnight, incubated for 16 hours with CDK4/6 inhibitor, and next incubated for 8 hours with CDK4/6 inhibitor and doxorubicin. Fresh media was then added, and cell number and viability were quantified 5 days later. γ-H2AX levels were assessed by flow cytometry after fixation and anti–γ-H2AX–FITC incubation (Millipore). Western blots were performed on cell lysates in NP-40 lysis buffer with protease inhibitors (Roche) and phosphatase inhibitors (Calbiochem) as previously described (11), using anti–p53-phospho-Ser15 (Cell Signaling), Bax, and actin-HRP (Santa Cruz Biotechnology Inc.).
Comet tail assay after IR. tHDFs received 6 Gy IR from a RS2000 Biological Irradiator (RadSource Inc.), with or without 24 hours of prior exposure to 100 nM PD0332991. Cells were placed on ice 5 minutes after IR and processed, as previously described (57), prior to electrophoresis with SYBR Green (Applied Biosystems), imaged as for γ-H2AX (see below), and quantified using Comet Score software (TriTek Co.).
γ-H2AX and clonogenic assay after IR. For γ-H2AX images, tHDFs received 6 Gy IR, with or without 24 hours of prior exposure to 100 nM PD0332991. Five minutes after IR, cell were washed twice with ice-cold PBS, fixed with 4% paraformaldehyde plus 0.1% Triton-X (Sigma-Aldrich) for 30 minutes, washed twice with ice-cold PBS, incubated with anti–γ-H2AX–Alexa Fluor 488 (Cell Signaling) and phalloidin–Alexa Fluor 568 (Invitrogen) for 1 hour, washed 4 times with ice-cold PBS, and mounted. Images were captured using a mercury laser attached to an inverted microscope (model IX-81, Olympus), which was equipped with a ×20 or a ×40 PlanApo objective and a CCD camera (model C4742-80-12AG, OCAR-ER, Hamamatsu), which was controlled by Slidebook software. Mean nuclear intensities were computed using ImageJ software ( http://rsbweb.nih.gov/ij/). The clonogenic assay was performed as previously described (58), with cells plated in a 6-well plate 6 hours prior to treatment for 24 hours with 100 nM PD0332991, with 6 Gy IR occurring 12 hours after drug exposure.
Compounds. R547 (17) was synthesized by the Center for Integrative Chemical Biology and Drug Discovery (CICBDD). 2BrIC was synthesized by OTAVA Chemicals, based on compound 4d, as shown in ref. 15. PD0332991 (13, 14) was provided by Pfizer Inc. or synthesized by the CICBDD. Flavopiridol (18) was purchased from Sigma-Aldrich. Roscovitine (16) was purchased from LC Laboratories.
BM-MNC isolation and CAFC assay. BM-MNC isolation and CAFC assays were performed as previously described (59). Briefly, bone marrow was harvested from femurs and tibias of mice and centrifuged to purify BM-MNCs. The frequencies of CAFCs were determined at weekly intervals (on day 7, 14, and 35). Wells were scored positive if at least 1 phase-dark hematopoietic clone (containing 5 or more cells) was seen. The frequency of CAFCs was then calculated using L-Calc software (StemCell Technologies) for limiting dilution analysis.
Bone marrow immunophenotyping and proliferation by flow cytometry. For HSPC proliferation experiments, mice received daily oral gavage with PD0332991 for 2 days, with an intraperitoneal injection of 1 mg BrdU every 6 hours for 24 hours prior to sacrifice. BM-MNC harvest and immunophenotyping was performed using rbc lysis, biotin-conjugated Lin-panel incubation (Invitrogen), paramagnetic bead-conjugated streptavidin (Miltenyi Biotec) incubation, and magnetic depletion, using an AutoMACS (Miltenyi Biotec). At least 2 × 106 Lin-depleted cells per mouse were incubated with fluorescently labeled antibodies against cell surface antigens used to identify hematopoietic progenitor subpopulations, as previously described (23, 60): CD34-FITC, CD16/32–Pacific Blue, IL7Ra-PE-Cy5, and cKit-APC–Alexa Fluor 750 from eBiosciences; Sca1-PE-Cy7, CD150-PE-Cy5, and CD48–Pacific Blue from BioLegend; and Aqua Live/Dead viability dye (Invitrogen). Streptavidin-PE–Texas Red (Invitrogen) was used to confirm efficiency of lineage depletion (data not shown). After cell surface staining, cells were fixed, permeabilized, and stained with antibodies against Ki67-FITC, BrdU-APC, and caspase3-PE (BD Biosciences). In all experiments, gating based on isotype controls was used as appropriate (data not shown). Flow cytometry was performed using a CyAn ADP (Dako) and analyzed with FlowJo software (Tree Star). For each bone marrow sample, a minimum of 200,000 cells were analyzed using FlowJo software (Tree Star). For cell culture samples, a minimum of 20,000 cells were analyzed.
Statistics. Unless otherwise noted, comparisons are made with 1-way ANOVA, with Bonferroni correction for multiple comparisons where appropriate. P values of less than 0.05 are considered significant. Error bars represent ± SEM.
-
Acknowledgments
We wish to thank Jim Bear, Arlene Bridges, Tao Cheng, Hanno Hock, Yan Liu, Chuck Perou, Gordon Peters, and Dave Roadcap for advice and reagents; Derrick Rossi, Sean Morrison, and Yue Xiong for critical reading of the manuscript; and Peter Toogood and Gerrit Los of Pfizer Inc., who supplied PD0332991 and compound-related expertise. This work was supported by the University of North Carolina Lineberger Comprehensive Cancer Center Mouse Phase I Unit and grants from the Golfers Against Cancer Foundation, the Ellison Medical Foundation, the Burroughs Wellcome Fund, and the National Institutes of Health (RO1-AG024379, RO1-AI077454, RO1-AI080432, T32-GM008719, F30-AG034806, GM008581). J.F. Bell was supported by a career development award from the Jimmy V foundation. K.B. Monahan was supported by an award from the National Cancer Foundation.
Address correspondence to: Norman E. Sharpless, The Lineberger Comprehensive Cancer Center, University of North Carolina School Medicine, CB #7295, Chapel Hill, North Carolina 27599, USA. Phone: 919.966.1185; Fax: 919.966.8212; E-mail: nes@med.unc.edu.
-
Footnotes
Conflict of interest: The University of North Carolina has filed a patent based on this work. Søren M. Johnson, Chad D. Torrice, Jessica F. Bell, Matthew R. Ramsey, and Norman E. Sharpless are co-inventors on this patent, which has been licensed to G-Zero Therapeutics, a company cofounded by Norman E. Sharpless and Kwok-Kin Wong.
Reference information: J Clin Invest. 2010;120(7):2528–2536. doi:10.1172/JCI41402.
See the related article at Radioprotection: smart games with death.
-
References
- Sinclair WK, Morton RA. X-ray sensitivity during cell generation cycle of cultured chinese hamster cells. Radiat Res. 1966;29(3):450–474.
- Terasima T, Tolmach LJ. X-ray sensitivity and DNA synthesis in synchronous populations of HeLa cells. Science. 1963;140:490–492.
- Little JB. Repair of sub-lethal and potentially lethal radiation damage in plateau phase cultures of human cells. Nature. 1969;224(5221):804–806.
- Elkind MM, Sutton H. X-ray damage and recovery in mammalian cells in culture. Nature. 1959;184:1293–1295.View this article via: PubMed Google Scholar
- Elkind MM, Sutton H. Radiation response of mammalian cells grown in culture. 1. Repair of x-ray damage in surviving chinese hamster cells. Radiat Res. 1960;13:556–593.
- Iliakis G, Wang Y, Guan J, Wang HC. DNA damage checkpoint control in cells exposed to ionizing radiation. Oncogene. 2003;22(37):5834–5847.
- Malumbres M. Revisiting the “Cdk-centric” view of the mammalian cell cycle. Cell Cycle. 2005;4(2):206–210.View this article via: PubMed Google Scholar
- Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18(22):2699–2711.
- Kozar K, et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 2004;118(4):477–491.
- Malumbres M, et al. Mammalian cells cycle without the D-type cyclin-elependent kinases Cdk4 and Cdk6. Cell. 2004;118(4):493–504.
- Ramsey MR, et al. Expression of p16(Ink4a) compensates for p18(Ink4c) loss in cyclin-dependent kinase 4/6-dependent tumors and tissues. Cancer Res. 2007;67(10):4732–4741.
- Shapiro GI. Cyclin-dependent kinase pathways as targets for cancer treatment. J Clin Oncol. 2006;24(11):1770–1783.View this article via: PubMed Google Scholar
- Fry DW, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther. 2004;3(11):1427–1437.View this article via: PubMed Google Scholar
- Toogood PL, et al. Discovery of a potent and selective inhibitor of cyclin-dependent kinase 4/6. J Med Chem. 2005;48(7):2388–2406.
- Zhu GX, et al. Synthesis, structure-activity relationship, and biological studies of indolocarbazoles as potent cyclin D1-CDK4 inhibitors. J Med Chem. 2003;46(11):2027–2030.
- Meijer L, et al. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur J Biochem. 1997;243(1–2):527–536.
- Chu XJ, et al. Discovery of [4-amino-2-(1-methanesulfonylpiperidin-4-ylamino) pyrimidin-5-yl](2,3-difluoro-6-methoxyphenyl)methanone (R547), a potent and selective cyclin-dependent kinase inhibitor with significant in vivo antitumor activity. J Med Chem. 2006;49(22):6549–6560.
- Kaur G, et al. Growth-inhibition with reversible cell-cycle arrest of carcinoma-cells by flavone-L86-8275. J Natl Cancer Inst. 1992;84(22):1736–1740.
- Johnson N, et al. Cdk1 participates in BRCA1-dependent S phase checkpoint control in response to DNA damage. Mol Cell. 2009;35(3):327–339.
- Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284(14):9558–9565.
- Chen X, et al. Protection of normal proliferating cells against chemotherapy by staurosporine-mediated, selective, and reversible G(1) arrest. J Natl Cancer Inst. 2000;92(24):1999–2008.
- Kiel MJ, et al. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449(7159):238–242.
- Passegue E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J Exp Med. 2005;202(11):1599–1611.
- O’Dwyer PJ, et al. A Phase I dose escalation trial of a daily oral CDK 4/6 inhibitor PD-0332991. J Clin Oncol. 2007;25(18S):3550.
- Vaughn DJ, et al. Treatment of growing teratoma syndrome. N Engl J Med. 2009;360(4):423–424.
- Burdelya LG, et al. An agonist of toll-like receptor 5 has radioprotective activity in mouse and primate models. Science. 2008;320(5873):226–230.
- Weiss JF, Landauer MR. History and development of radiation-protective agents. Int J Radiat Biol. 2009;85(7):539–573.
- Nakorn TN, Traver D, Weissman IL, Akashi K. Myeloerythroid-restricted progenitors are sufficient to confer radioprotection and provide the majority of day 8 CFU-S. J Clin Invest. 2002;109(12):1579–1585.
- Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. P21 is a universal inhibitor of cyclin kinases. Nature. 1993;366(6456):701–704.
- Dulic V, et al. P53-dependent inhibition of cyclin-dependent kinase-activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76(6):1013–1023.View this article via: PubMed Google Scholar
- Cheng T, et al. Hematopoietic stem cell quiescence maintained by p21(cip1/waf1). Science. 2000;287(5459):1804–1808.View this article via: PubMed Google Scholar
- Herodin F, Bourin P, Mayol JF, Lataillade JJ, Drouet M. Short-term injection of antiapoptotic cytokine combinations soon after lethal gamma-irradiation promotes survival. Blood. 2003;101(7):2609–2616.
- Chin L, et al. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 1997;11(21):2822–2834.
- Sharpless NE, Kannan K, Xu J, Bosenberg MW, Chin LD. Both products of the mouse Ink4a/Arf locus suppress melanoma formation in vivo. Oncogene. 2003;22(32):5055–5059.
- Rothe WE, Grenan MM. Radioprotection by mitotic inhibitors and mercaptoethylamine. Science. 1961;133:888.
- Smith WW. Protective effect of a colchicine derivative in mice exposed to x-radiation. Science. 1958;127(3294):340–341.View this article via: PubMed Google Scholar
- Deans AJ, Khanna KK, McNees CJ, Mercurio C, Heierhorst J, McArthur GA. Cyclin-dependent kinase 2 functions in normal DNA repair and is a therapeutic target in BRCA1-deficient cancers. Cancer Res. 2006;66(16):8219–8226.
- Maude SL, Enders GH. Cdk inhibition in human cells compromises Chk1 function and activates a DNA damage response. Cancer Res. 2005;65(3):780–786.View this article via: PubMed Google Scholar
- Mosesso P, et al. Relationship between chromatin structure, DNA damage and repair following X-irradiation of human lymphocytes [published online ahead of print March 16, 2010]. Mutat Res. doi:10.1016/j.mrgentox.2010.03.005.
- Sun Y, Jiang X, Price BD. Tip60: Connecting chromatin to DNA damage signaling. Cell Cycle. 2010;9(5):930–936.View this article via: PubMed Google Scholar
- Iliakis G, Nusse M. The importance of G1/S-border and mitosis in the fixation of potentially lethal damage. Radiat Environ Biophys. 1983;22(3):201–207.
- Phillips AC, Bates S, Ryan KM, Helin K, Vousden KH. Induction of DNA synthesis and apoptosis are separable functions of E2F-1. Genes Dev. 1997;11(14):1853–1863.View this article via: PubMed Google Scholar
- Tyagi S, Herr W. E2F1 mediates DNA damage and apoptosis through HCF-1 and the MLL family of histone methyltransferases. EMBO J. 2009;28(20):3185–3195.View this article via: PubMed Google Scholar
- Pardee AB. Restriction point for control of normal animal-cell proliferation. Proc Natl Acad Sci U S A. 1974;71(4):1286–1290.
- Blagosklonny MV, Pardee AB. The restriction point of the cell cycle. Cell Cycle. 2002;1(2):103–110.View this article via: PubMed Google Scholar
- Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9(8):619–631.
- Roots R, Kraft G, Gosschalk E. The formation of radiation-induced DNA breaks - the ratio of double-strand breaks to single-strand breaks. Int J Radiat Oncol Biol Phys. 1985;11(2):259–265.View this article via: PubMed Google Scholar
- Dextraze ME, Gantchev T, Girouard S, Hunting D. . DNA interstrand cross–links induced by ionizing radiation: An unsung lesion [published online ahead of print January 15, 2010]. Mutat Res. doi:10.1016/j.mrrev.2009.12.007.
- Dileonardo A, Linke SP, Clarkin K, Wahl GM. DNA-damage triggers a prolonged P53-dependent G(1) arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 1994;8(21):2540–2551.
- Santra MK, Wajapeyee N, Green MR. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459(7247):722–725.View this article via: PubMed Google Scholar
- Uckun FM, Souza L, Waddick KG, Wick M, Song CW. In vivo radioprotective effects of recombinant human granulocyte colony-stimulating factor in lethally irradiated mice. Blood. 1990;75(3):638–645.View this article via: PubMed Google Scholar
- Hershman D, et al. Acute myeloid leukemia or myelodysplastic syndrome following use of granulocyte colony-stimulating factors during breast cancer adjuvant chemotherapy. J Natl Cancer Inst. 2007;99(3):196–205.View this article via: PubMed Google Scholar
- Le Deley MC, et al. Anthracyclines, mitoxantrone, radiotherapy, and granulocyte colony-stimulating factor: Risk factors for leukemia and myelodysplastic syndrome after breast cancer. J Clin Oncol. 2007;25(3):292–300.View this article via: PubMed Google Scholar
- Pevarello P, Villa M. Cyclin-dependent kinase inhibitors: a survey of the recent patent literature. Expert Opin Ther Pat. 2005;15(6):675–703.View this article via: CrossRef Google Scholar
- Wang YA, Elson A, Leder P. Loss of p21 increases sensitivity to ionizing radiation and delays the onset of lymphoma in atm-deficient mice. Proc Natl Acad Sci U S A. 1997;94(26):14590–14595.
- Shields JM, et al. Lack of extracellular signal-regulated kinase mitogen-activated protein kinase signaling shows a new type of melanoma. Cancer Res. 2007;67(4):1502–1512.
- Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc. 2006;1(1):23–29.
- Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1(5):2315–2319.
- Meng AM, Wang Y, Van Zant G, Zhou DH. Ionizing radiation and busulfan induce premature senescence in murine bone marrow hematopoietic cells. Cancer Res. 2003;63(17):5414–5419.View this article via: PubMed Google Scholar
- Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121(7):1109–1121.
-
Version history
- Version 1 (June 23, 2010): No description
- Version 2 (July 1, 2010): No description



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)






