Advertisement
Research Article Free access | 10.1172/JCI39060
Homozygous loss-of-function mutations in the gene encoding the dopamine transporter are associated with infantile parkinsonism-dystonia
Manju A. Kurian,1,2 Juan Zhen,3 Shu-Yuan Cheng,3 Yan Li,3 Santosh R. Mordekar,4 Philip Jardine,5 Neil V. Morgan,1 Esther Meyer,1 Louise Tee,1 Shanaz Pasha,1 Evangeline Wassmer,2 Simon J.R. Heales,6 Paul Gissen,1 Maarten E.A. Reith,3,7 and Eamonn R. Maher1,8
1Department of Medical and Molecular Genetics, University of Birmingham School of Medicine, Institute of Biomedical Research, Birmingham, United Kingdom. 2Department of Paediatric Neurology, Birmingham Children’s Hospital, Birmingham, United Kingdom. 3Department of Psychiatry, Millhauser Laboratories, New York University School of Medicine, New York, New York, USA. 4Department of Paediatric Neurology, Sheffield Children’s Hospital, Sheffield, United Kingdom. 5Department of Paediatric Neurology, Bristol Children’s Hospital, Bristol, United Kingdom. 6Neurometabolic Unit, National Hospital, and Department of Chemical Pathology, Great Ormond Street Hospital, London, United Kingdom. 7Department of Pharmacology, New York University School of Medicine, New York, New York, USA. 8West Midlands Regional Genetics Service, Birmingham Women’s Hospital, Birmingham, United Kingdom.
Address correspondence to: Eamonn R. Maher, Department of Medical and Molecular Genetics, Institute of Biomedical Research, Birmingham University, Edgbaston, Birmingham B15 2TT, United Kingdom. Phone: 00-44-121-627-2741; Fax: 00-44-121-414-2538; E-mail: e.r.maher@bham.ac.uk.
Find articles by Kurian, M. in: PubMed | Google Scholar
1Department of Medical and Molecular Genetics, University of Birmingham School of Medicine, Institute of Biomedical Research, Birmingham, United Kingdom. 2Department of Paediatric Neurology, Birmingham Children’s Hospital, Birmingham, United Kingdom. 3Department of Psychiatry, Millhauser Laboratories, New York University School of Medicine, New York, New York, USA. 4Department of Paediatric Neurology, Sheffield Children’s Hospital, Sheffield, United Kingdom. 5Department of Paediatric Neurology, Bristol Children’s Hospital, Bristol, United Kingdom. 6Neurometabolic Unit, National Hospital, and Department of Chemical Pathology, Great Ormond Street Hospital, London, United Kingdom. 7Department of Pharmacology, New York University School of Medicine, New York, New York, USA. 8West Midlands Regional Genetics Service, Birmingham Women’s Hospital, Birmingham, United Kingdom.
Address correspondence to: Eamonn R. Maher, Department of Medical and Molecular Genetics, Institute of Biomedical Research, Birmingham University, Edgbaston, Birmingham B15 2TT, United Kingdom. Phone: 00-44-121-627-2741; Fax: 00-44-121-414-2538; E-mail: e.r.maher@bham.ac.uk.
Find articles by Zhen, J. in: PubMed | Google Scholar
1Department of Medical and Molecular Genetics, University of Birmingham School of Medicine, Institute of Biomedical Research, Birmingham, United Kingdom. 2Department of Paediatric Neurology, Birmingham Children’s Hospital, Birmingham, United Kingdom. 3Department of Psychiatry, Millhauser Laboratories, New York University School of Medicine, New York, New York, USA. 4Department of Paediatric Neurology, Sheffield Children’s Hospital, Sheffield, United Kingdom. 5Department of Paediatric Neurology, Bristol Children’s Hospital, Bristol, United Kingdom. 6Neurometabolic Unit, National Hospital, and Department of Chemical Pathology, Great Ormond Street Hospital, London, United Kingdom. 7Department of Pharmacology, New York University School of Medicine, New York, New York, USA. 8West Midlands Regional Genetics Service, Birmingham Women’s Hospital, Birmingham, United Kingdom.
Address correspondence to: Eamonn R. Maher, Department of Medical and Molecular Genetics, Institute of Biomedical Research, Birmingham University, Edgbaston, Birmingham B15 2TT, United Kingdom. Phone: 00-44-121-627-2741; Fax: 00-44-121-414-2538; E-mail: e.r.maher@bham.ac.uk.
Find articles by Cheng, S. in: PubMed | Google Scholar
1Department of Medical and Molecular Genetics, University of Birmingham School of Medicine, Institute of Biomedical Research, Birmingham, United Kingdom. 2Department of Paediatric Neurology, Birmingham Children’s Hospital, Birmingham, United Kingdom. 3Department of Psychiatry, Millhauser Laboratories, New York University School of Medicine, New York, New York, USA. 4Department of Paediatric Neurology, Sheffield Children’s Hospital, Sheffield, United Kingdom. 5Department of Paediatric Neurology, Bristol Children’s Hospital, Bristol, United Kingdom. 6Neurometabolic Unit, National Hospital, and Department of Chemical Pathology, Great Ormond Street Hospital, London, United Kingdom. 7Department of Pharmacology, New York University School of Medicine, New York, New York, USA. 8West Midlands Regional Genetics Service, Birmingham Women’s Hospital, Birmingham, United Kingdom.
Address correspondence to: Eamonn R. Maher, Department of Medical and Molecular Genetics, Institute of Biomedical Research, Birmingham University, Edgbaston, Birmingham B15 2TT, United Kingdom. Phone: 00-44-121-627-2741; Fax: 00-44-121-414-2538; E-mail: e.r.maher@bham.ac.uk.
Find articles by Li, Y. in: PubMed | Google Scholar
1Department of Medical and Molecular Genetics, University of Birmingham School of Medicine, Institute of Biomedical Research, Birmingham, United Kingdom. 2Department of Paediatric Neurology, Birmingham Children’s Hospital, Birmingham, United Kingdom. 3Department of Psychiatry, Millhauser Laboratories, New York University School of Medicine, New York, New York, USA. 4Department of Paediatric Neurology, Sheffield Children’s Hospital, Sheffield, United Kingdom. 5Department of Paediatric Neurology, Bristol Children’s Hospital, Bristol, United Kingdom. 6Neurometabolic Unit, National Hospital, and Department of Chemical Pathology, Great Ormond Street Hospital, London, United Kingdom. 7Department of Pharmacology, New York University School of Medicine, New York, New York, USA. 8West Midlands Regional Genetics Service, Birmingham Women’s Hospital, Birmingham, United Kingdom.
Address correspondence to: Eamonn R. Maher, Department of Medical and Molecular Genetics, Institute of Biomedical Research, Birmingham University, Edgbaston, Birmingham B15 2TT, United Kingdom. Phone: 00-44-121-627-2741; Fax: 00-44-121-414-2538; E-mail: e.r.maher@bham.ac.uk.
Find articles by Mordekar, S. in: PubMed | Google Scholar
1Department of Medical and Molecular Genetics, University of Birmingham School of Medicine, Institute of Biomedical Research, Birmingham, United Kingdom. 2Department of Paediatric Neurology, Birmingham Children’s Hospital, Birmingham, United Kingdom. 3Department of Psychiatry, Millhauser Laboratories, New York University School of Medicine, New York, New York, USA. 4Department of Paediatric Neurology, Sheffield Children’s Hospital, Sheffield, United Kingdom. 5Department of Paediatric Neurology, Bristol Children’s Hospital, Bristol, United Kingdom. 6Neurometabolic Unit, National Hospital, and Department of Chemical Pathology, Great Ormond Street Hospital, London, United Kingdom. 7Department of Pharmacology, New York University School of Medicine, New York, New York, USA. 8West Midlands Regional Genetics Service, Birmingham Women’s Hospital, Birmingham, United Kingdom.
Address correspondence to: Eamonn R. Maher, Department of Medical and Molecular Genetics, Institute of Biomedical Research, Birmingham University, Edgbaston, Birmingham B15 2TT, United Kingdom. Phone: 00-44-121-627-2741; Fax: 00-44-121-414-2538; E-mail: e.r.maher@bham.ac.uk.
Find articles by Jardine, P. in: PubMed | Google Scholar
1Department of Medical and Molecular Genetics, University of Birmingham School of Medicine, Institute of Biomedical Research, Birmingham, United Kingdom. 2Department of Paediatric Neurology, Birmingham Children’s Hospital, Birmingham, United Kingdom. 3Department of Psychiatry, Millhauser Laboratories, New York University School of Medicine, New York, New York, USA. 4Department of Paediatric Neurology, Sheffield Children’s Hospital, Sheffield, United Kingdom. 5Department of Paediatric Neurology, Bristol Children’s Hospital, Bristol, United Kingdom. 6Neurometabolic Unit, National Hospital, and Department of Chemical Pathology, Great Ormond Street Hospital, London, United Kingdom. 7Department of Pharmacology, New York University School of Medicine, New York, New York, USA. 8West Midlands Regional Genetics Service, Birmingham Women’s Hospital, Birmingham, United Kingdom.
Address correspondence to: Eamonn R. Maher, Department of Medical and Molecular Genetics, Institute of Biomedical Research, Birmingham University, Edgbaston, Birmingham B15 2TT, United Kingdom. Phone: 00-44-121-627-2741; Fax: 00-44-121-414-2538; E-mail: e.r.maher@bham.ac.uk.
Find articles by Morgan, N. in: PubMed | Google Scholar
1Department of Medical and Molecular Genetics, University of Birmingham School of Medicine, Institute of Biomedical Research, Birmingham, United Kingdom. 2Department of Paediatric Neurology, Birmingham Children’s Hospital, Birmingham, United Kingdom. 3Department of Psychiatry, Millhauser Laboratories, New York University School of Medicine, New York, New York, USA. 4Department of Paediatric Neurology, Sheffield Children’s Hospital, Sheffield, United Kingdom. 5Department of Paediatric Neurology, Bristol Children’s Hospital, Bristol, United Kingdom. 6Neurometabolic Unit, National Hospital, and Department of Chemical Pathology, Great Ormond Street Hospital, London, United Kingdom. 7Department of Pharmacology, New York University School of Medicine, New York, New York, USA. 8West Midlands Regional Genetics Service, Birmingham Women’s Hospital, Birmingham, United Kingdom.
Address correspondence to: Eamonn R. Maher, Department of Medical and Molecular Genetics, Institute of Biomedical Research, Birmingham University, Edgbaston, Birmingham B15 2TT, United Kingdom. Phone: 00-44-121-627-2741; Fax: 00-44-121-414-2538; E-mail: e.r.maher@bham.ac.uk.
Find articles by Meyer, E. in: PubMed | Google Scholar
1Department of Medical and Molecular Genetics, University of Birmingham School of Medicine, Institute of Biomedical Research, Birmingham, United Kingdom. 2Department of Paediatric Neurology, Birmingham Children’s Hospital, Birmingham, United Kingdom. 3Department of Psychiatry, Millhauser Laboratories, New York University School of Medicine, New York, New York, USA. 4Department of Paediatric Neurology, Sheffield Children’s Hospital, Sheffield, United Kingdom. 5Department of Paediatric Neurology, Bristol Children’s Hospital, Bristol, United Kingdom. 6Neurometabolic Unit, National Hospital, and Department of Chemical Pathology, Great Ormond Street Hospital, London, United Kingdom. 7Department of Pharmacology, New York University School of Medicine, New York, New York, USA. 8West Midlands Regional Genetics Service, Birmingham Women’s Hospital, Birmingham, United Kingdom.
Address correspondence to: Eamonn R. Maher, Department of Medical and Molecular Genetics, Institute of Biomedical Research, Birmingham University, Edgbaston, Birmingham B15 2TT, United Kingdom. Phone: 00-44-121-627-2741; Fax: 00-44-121-414-2538; E-mail: e.r.maher@bham.ac.uk.
Find articles by Tee, L. in: PubMed | Google Scholar
1Department of Medical and Molecular Genetics, University of Birmingham School of Medicine, Institute of Biomedical Research, Birmingham, United Kingdom. 2Department of Paediatric Neurology, Birmingham Children’s Hospital, Birmingham, United Kingdom. 3Department of Psychiatry, Millhauser Laboratories, New York University School of Medicine, New York, New York, USA. 4Department of Paediatric Neurology, Sheffield Children’s Hospital, Sheffield, United Kingdom. 5Department of Paediatric Neurology, Bristol Children’s Hospital, Bristol, United Kingdom. 6Neurometabolic Unit, National Hospital, and Department of Chemical Pathology, Great Ormond Street Hospital, London, United Kingdom. 7Department of Pharmacology, New York University School of Medicine, New York, New York, USA. 8West Midlands Regional Genetics Service, Birmingham Women’s Hospital, Birmingham, United Kingdom.
Address correspondence to: Eamonn R. Maher, Department of Medical and Molecular Genetics, Institute of Biomedical Research, Birmingham University, Edgbaston, Birmingham B15 2TT, United Kingdom. Phone: 00-44-121-627-2741; Fax: 00-44-121-414-2538; E-mail: e.r.maher@bham.ac.uk.
Find articles by Pasha, S. in: PubMed | Google Scholar
1Department of Medical and Molecular Genetics, University of Birmingham School of Medicine, Institute of Biomedical Research, Birmingham, United Kingdom. 2Department of Paediatric Neurology, Birmingham Children’s Hospital, Birmingham, United Kingdom. 3Department of Psychiatry, Millhauser Laboratories, New York University School of Medicine, New York, New York, USA. 4Department of Paediatric Neurology, Sheffield Children’s Hospital, Sheffield, United Kingdom. 5Department of Paediatric Neurology, Bristol Children’s Hospital, Bristol, United Kingdom. 6Neurometabolic Unit, National Hospital, and Department of Chemical Pathology, Great Ormond Street Hospital, London, United Kingdom. 7Department of Pharmacology, New York University School of Medicine, New York, New York, USA. 8West Midlands Regional Genetics Service, Birmingham Women’s Hospital, Birmingham, United Kingdom.
Address correspondence to: Eamonn R. Maher, Department of Medical and Molecular Genetics, Institute of Biomedical Research, Birmingham University, Edgbaston, Birmingham B15 2TT, United Kingdom. Phone: 00-44-121-627-2741; Fax: 00-44-121-414-2538; E-mail: e.r.maher@bham.ac.uk.
Find articles by Wassmer, E. in: PubMed | Google Scholar
1Department of Medical and Molecular Genetics, University of Birmingham School of Medicine, Institute of Biomedical Research, Birmingham, United Kingdom. 2Department of Paediatric Neurology, Birmingham Children’s Hospital, Birmingham, United Kingdom. 3Department of Psychiatry, Millhauser Laboratories, New York University School of Medicine, New York, New York, USA. 4Department of Paediatric Neurology, Sheffield Children’s Hospital, Sheffield, United Kingdom. 5Department of Paediatric Neurology, Bristol Children’s Hospital, Bristol, United Kingdom. 6Neurometabolic Unit, National Hospital, and Department of Chemical Pathology, Great Ormond Street Hospital, London, United Kingdom. 7Department of Pharmacology, New York University School of Medicine, New York, New York, USA. 8West Midlands Regional Genetics Service, Birmingham Women’s Hospital, Birmingham, United Kingdom.
Address correspondence to: Eamonn R. Maher, Department of Medical and Molecular Genetics, Institute of Biomedical Research, Birmingham University, Edgbaston, Birmingham B15 2TT, United Kingdom. Phone: 00-44-121-627-2741; Fax: 00-44-121-414-2538; E-mail: e.r.maher@bham.ac.uk.
Find articles by Heales, S. in: PubMed | Google Scholar
1Department of Medical and Molecular Genetics, University of Birmingham School of Medicine, Institute of Biomedical Research, Birmingham, United Kingdom. 2Department of Paediatric Neurology, Birmingham Children’s Hospital, Birmingham, United Kingdom. 3Department of Psychiatry, Millhauser Laboratories, New York University School of Medicine, New York, New York, USA. 4Department of Paediatric Neurology, Sheffield Children’s Hospital, Sheffield, United Kingdom. 5Department of Paediatric Neurology, Bristol Children’s Hospital, Bristol, United Kingdom. 6Neurometabolic Unit, National Hospital, and Department of Chemical Pathology, Great Ormond Street Hospital, London, United Kingdom. 7Department of Pharmacology, New York University School of Medicine, New York, New York, USA. 8West Midlands Regional Genetics Service, Birmingham Women’s Hospital, Birmingham, United Kingdom.
Address correspondence to: Eamonn R. Maher, Department of Medical and Molecular Genetics, Institute of Biomedical Research, Birmingham University, Edgbaston, Birmingham B15 2TT, United Kingdom. Phone: 00-44-121-627-2741; Fax: 00-44-121-414-2538; E-mail: e.r.maher@bham.ac.uk.
Find articles by Gissen, P. in: PubMed | Google Scholar
1Department of Medical and Molecular Genetics, University of Birmingham School of Medicine, Institute of Biomedical Research, Birmingham, United Kingdom. 2Department of Paediatric Neurology, Birmingham Children’s Hospital, Birmingham, United Kingdom. 3Department of Psychiatry, Millhauser Laboratories, New York University School of Medicine, New York, New York, USA. 4Department of Paediatric Neurology, Sheffield Children’s Hospital, Sheffield, United Kingdom. 5Department of Paediatric Neurology, Bristol Children’s Hospital, Bristol, United Kingdom. 6Neurometabolic Unit, National Hospital, and Department of Chemical Pathology, Great Ormond Street Hospital, London, United Kingdom. 7Department of Pharmacology, New York University School of Medicine, New York, New York, USA. 8West Midlands Regional Genetics Service, Birmingham Women’s Hospital, Birmingham, United Kingdom.
Address correspondence to: Eamonn R. Maher, Department of Medical and Molecular Genetics, Institute of Biomedical Research, Birmingham University, Edgbaston, Birmingham B15 2TT, United Kingdom. Phone: 00-44-121-627-2741; Fax: 00-44-121-414-2538; E-mail: e.r.maher@bham.ac.uk.
Find articles by Reith, M. in: PubMed | Google Scholar
1Department of Medical and Molecular Genetics, University of Birmingham School of Medicine, Institute of Biomedical Research, Birmingham, United Kingdom. 2Department of Paediatric Neurology, Birmingham Children’s Hospital, Birmingham, United Kingdom. 3Department of Psychiatry, Millhauser Laboratories, New York University School of Medicine, New York, New York, USA. 4Department of Paediatric Neurology, Sheffield Children’s Hospital, Sheffield, United Kingdom. 5Department of Paediatric Neurology, Bristol Children’s Hospital, Bristol, United Kingdom. 6Neurometabolic Unit, National Hospital, and Department of Chemical Pathology, Great Ormond Street Hospital, London, United Kingdom. 7Department of Pharmacology, New York University School of Medicine, New York, New York, USA. 8West Midlands Regional Genetics Service, Birmingham Women’s Hospital, Birmingham, United Kingdom.
Address correspondence to: Eamonn R. Maher, Department of Medical and Molecular Genetics, Institute of Biomedical Research, Birmingham University, Edgbaston, Birmingham B15 2TT, United Kingdom. Phone: 00-44-121-627-2741; Fax: 00-44-121-414-2538; E-mail: e.r.maher@bham.ac.uk.
Find articles by Maher, E. in: PubMed | Google Scholar
Published May 26, 2009 - More info
J Clin Invest. 2009;119(6):1595–1603. https://doi.org/10.1172/JCI39060.
© 2009 The American Society for Clinical Investigation
Received: February 27, 2009; Accepted: April 8, 2009
Related article:
Abstract
The dopamine transporter (DAT) retrieves the neurotransmitter dopamine from the synaptic cleft at dopaminergic synapses. Variations in solute carrier family 6A, member 3 (SLC6A3/DAT1), the human gene encoding DAT, have been implicated in attention deficit hyperactivity and bipolar disorders, and DAT is a prominent site of action for drugs such as amphetamines and cocaine. In this issue of the JCI, Kurian et al. report that an autosomal recessive infantile parkinsonism-dystonia is caused by loss-of-function mutations in DAT that impair dopamine reuptake (see the related article beginning on page 1595). Though this might be predicted to result in dopamine excess in the synaptic cleft, it likely also causes depletion of presynaptic dopamine stores and possibly downregulation of postsynaptic dopamine receptor function, resulting in impairments in dopaminergic neurotransmission consistent with the clinical presentation. This is the first report of a genetic alteration in DAT function underlying a parkinsonian disorder.
Authors
Craig Blackstone
-
Abstract
Genetic variants of the SLC6A3 gene that encodes the human dopamine transporter (DAT) have been linked to a variety of neuropsychiatric disorders, particularly attention deficit hyperactivity disorder. In addition, the homozygous Slc6a3 knockout mouse displays a hyperactivity phenotype. Here, we analyzed 2 unrelated consanguineous families with infantile parkinsonism-dystonia (IPD) syndrome and identified homozygous missense SLC6A3 mutations (p.L368Q and p.P395L) in both families. Functional studies demonstrated that both mutations were loss-of-function mutations that severely reduced levels of mature (85-kDa) DAT while having a differential effect on the apparent binding affinity of dopamine. Thus, in humans, loss-of-function SLC6A3 mutations that impair DAT-mediated dopamine transport activity are associated with an early-onset complex movement disorder. Identification of the molecular basis of IPD suggests SLC6A3 as a candidate susceptibility gene for other movement disorders associated with parkinsonism and/or dystonic features.
-
Introduction
Parkinson disease is the second most common neurodegenerative disorder after Alzheimer disease and affects approximately 1% of the population over 50 years of age. Although most cases of Parkinson disease are sporadic, a number of inherited disorders can be associated with parkinsonism. Molecular genetic investigation of these cases can provide insights into the pathogenesis of movement disorders and/or neurodegeneration. The subgroup of patients who present with early-onset disease are of particular interest. Infantile parkinsonism-dystonia (IPD) is a severe neurological syndrome that usually presents in early infancy with hypokinetic parkinsonism (1). Subsequently, a complex movement disorder with dystonia develops in association with axial hypotonia and limb hypertonicity. The condition is severe and can be fatal. IPD is rare but is often underdiagnosed, as the clinical presentation can mimic certain types of cerebral palsy. Elucidating the molecular basis of IPD might provide insights into disease pathogenesis and allow earlier diagnosis.
In order to define the molecular basis of IPD, we used autozygosity mapping techniques to perform molecular genetic investigations in 2 consanguineous families. The disease locus was mapped to chromosome 5p15.3. We demonstrated different germline SLC6A3 gene mutations in both kindreds. Functional studies revealed that both mutations led to a reduction in the level of mature dopamine transporter (DAT).
-
Results
Two consanguineous families with IPD were investigated. Family 1 was a large consanguineous kindred of Pakistani origin (Figure 1A) with 2 affected children (patients 1 and 2), who were first cousins. Family 2 (Figure 1B) was of mixed European descent with 1 affected child (patient 3). The diagnosis of IPD was based on characteristic clinical features and cerebrospinal fluid (CSF) neurotransmitter studies (Table 1). In brief, following a normal pregnancy and birth, all children had neonatal irritability and feeding difficulties. Parkinsonism-dystonia heralded disease onset in early infancy and was rapidly followed by the development of pyramidal tract features. Two patients (patients 1 and 3) were initially misdiagnosed with cerebral palsy. On examination at ages 6–12 months, all patients had features of parkinsonism, dystonia, and pyramidal tract signs with evidence of global developmental delay. CSF biogenic amine metabolite studies in affected patients revealed markedly elevated concentrations of homovanillic acid (HVA; with normal 5-hydroxyindoleacetic acid levels). MRI brain scans did not reveal structural abnormalities in any of the patients. Formal neuropsychiatric assessments to determine whether the children showed any evidence of cognitive impairment were not conducted. All patients showed a poor clinical response to multiple therapeutic agents. CSF HVA levels did not normalize after L-dopa or tetrabenazine therapy. Insertion of a deep-brain stimulator device in patient 3 resulted in some symptomatic improvement of dystonia and rigidity. All children showed a progressive disease course with worsening symptoms of parkinsonism, dystonia, and hypertonicity. None of the affected children showed evidence of psychiatric or behavioral/conduct disorders. Detailed neurological examination of all the children’s parents (ages 21 to 37 years) and siblings (ages 1 to 9 years) revealed no neurological abnormalities. Additionally, none of the unaffected family members (including siblings, obligate carrier parents, parental siblings, and grandparents) were known to have a neuropsychiatric or age-related movement disorder.
 Figure 1
Figure 1Family trees of consanguineous kindreds. (A) Family 1 and (B) family 2 kinship. Children affected with IPD are indicated by black shading. Circles indicate females; squares indicate males; diamonds indicate undisclosed gender.
In the light of the diagnosis of autosomal recessive IPD and parental consanguinity, we undertook an autozygosity mapping study in family 1. Initially, genome-wide SNP genotyping (using the Affymetrix 250K SNP microarray) was performed in patients 1 and 2 from family 1. In these 2 affected cousins, 4 regions of extended homozygosity (>2 Mb) were detected (on chromosomes 5 [2.3 Mb], 7 [6 Mb], 9 [41 Mb], and 14 [9 Mb]). These regions were then further analyzed using polymorphic microsatellite markers (~0.5–2 Mb apart) mapping to these regions of interest. Linkage to the candidate regions on chromosomes 7 (4 markers), 9 (6 markers), and 14 (5 markers) was either excluded by segregation of the microsatellite marker alleles or the finding of heterozygous alleles in affected individuals (data not shown). On chromosome 5, the 2 children shared a common homozygous haplotype of 201 SNPs from 1 to 2,373,003 bp, thereby defining the margins of the homozygous region, while the unaffected sibling of patient 1 demonstrated SNP heterozygosity in this region (Figure 2). Genotyping of the family with 4 microsatellite markers (D5S2488, D5S1981, D5S2005, and D5678) within the interval revealed an identical homozygous haplotype in the 2 affected children, consistent with linkage to this region (Figure 3). Subsequent microsatellite marker analysis of family 2 was also consistent with disease linkage to this region (Figure 4). Multipoint linkage analysis gave a maximum lod score of 4.76 (family 1, 3.29; family 2, 1.46).
 Figure 2
Figure 2Localization of the disease locus on chromosome 5 on a 250K Affymetrix SNP array. SNP array data for patient 1 (VII:1) and patient 2 (VII:3) and the unaffected sibling of patient 1 (VII:2). Patients 1 and 2 displayed a region of SNP homozygosity from 0–2.3 Mb. The unaffected sibling did not display this common SNP homozygosity. Dark blue, AA homozygous SNP call; mid-blue, BB homozygous SNP call; light blue, no call; white, AB heterozygous call.
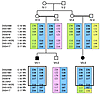 Figure 3
Figure 3Microsatellite marker linkage analysis on chromosome 5 in family 1. Affected individuals are indicated by black symbols. SLC6A3 gene is located at 5p15.3. Microsatellite markers are positioned according to physical distance (measured in Mb). Haplotypes for these markers are shown, and the disease-associated haplotypes are boxed in blue. Green, pink, and yellow boxes indicate non–disease-associated haplotypes. The 2 affected children shared a common homozygous haplotype in this region. N, novel microsatellite marker.
 Figure 4
Figure 4Microsatellite marker analysis on chromosome 5 in family 2. The black symbol indicates the affected patient. Microsatellite markers are positioned according to physical distance (measured in Mb). Haplotypes for these markers are shown, and the disease-associated haplotypes are boxed in red. Orange and blue boxes indicate non–disease-associated haplotypes. Microsatellite markers show evidence of segregation with disease status in this family.
After establishing the region on chromosome 5 as the location of the IPD gene, we examined the 39 genes within the region for a candidate IPD gene. SLC6A3 (located at chromosome 5; 1,445,909–1,498,538 bp) encoded a DAT, and mutation analysis of SLC6A3 was initiated. A homozygous putative missense mutation c.1103T>A (p.L368Q) (Figure 5) was detected at exon 8 in patients 1 and 2. Following identification of the putative mutation, SLC6A3 mutation testing was performed in family 2, and a homozygous putative mutation in exon 9, c.1184C>T (p.P395L), was detected in patient 3 (Figure 5). Both mutations segregated with IPD disease status in the families, and neither mutation was detected in an extensive analysis of ethnically matched control chromosomes (544 alleles from Asian individuals and 438 alleles from individuals of mixed European descent). Sequence alignment showed Leu368 and Pro395 to be highly conserved throughout the species (Figure 6). Both residues (particularly Leu368) were largely conserved in other human SLC6 transporters, suggesting that they are important for transporter function but perhaps not for dopamine-specific transporter properties.
 Figure 5
Figure 5SLC6A3 mutations in patients 1 and 3. The alignments of SLC6A3 nucleotides c.1091–c.1116 (A–C) and c.1168–c.1197 (D–F) are shown. B illustrates the c.1103T>A mutation in family 1. E illustrates the c.1184C>T mutation in family 2.
 Figure 6
Figure 6Conservation of the mutated DAT1 residues L368 and P395. Five representative vertebrate sequences are aligned. L368 and P395 are shown in red. Residues matching the consensus sequence are in gray; those not matching the consensus sequence are in white.
To determine the effects of the L368Q and P395L mutations, mutant human DAT (hDAT) proteins were transiently expressed in HEK293 cells and their transport activity compared with that of wild-type hDAT (see Methods and Table 2) (2–4). In experiments carried out in parallel, wild-type hDAT showed normal transport activity, whereas L368Q and P395L hDAT were devoid of uptake activity. The binding affinity of the cocaine analog [3H]2β-carbomethoxy-3β-(4-fluorophenyl)-tropane ([3H]CFT) was near normal in the mutants (30–36 nM, compared with 16 nM in wild-type hDAT; see Table 2). In contrast, the potency of dopamine in inhibiting cocaine analog binding was greatly reduced in L368Q hDAT (KI increased by an order of magnitude), whereas the potency of dopamine in P395L hDAT was close to that of wild type (1). Thus, both mutations were loss-of-function mutations with respect to the capability of DAT to translocate its substrate, dopamine, in conjunction with a loss of apparent binding affinity of dopamine in the case of one of the mutations. Maximal binding of [3H]CFT to cells, primarily representing surface binding (5), indicated appreciable expression of DAT in both mutants, although L368Q showed a reduction that was statistically significant (Table 2).
To investigate whether the observed reductions in dopamine transport were related to disordered production, location, or function of the mutant proteins, immunoblotting was performed. Analysis of whole cell lysates demonstrated equivalent levels of immature (55-kDa) DAT for wild-type and P395L, but there was some reduction for L368Q (total protein loading, as judged by β-actin, was equivalent for wild-type, L368Q, and P395L hDAT). However both mutant proteins demonstrated a profound reduction in mature (85-kDa) DAT compared with wild-type DAT. Analysis of surface (biotinylated DAT) showed a similar pattern (Figure 7).
 Figure 7
Figure 7Expression of the L368Q hDAT and P395L hDAT in transiently transfected HEK293 cells via immunoblotting analysis. Cells were transiently transfected with wild-type, L368Q, and P395L hDAT and reagent only. In each panel, the same amount of protein was loaded in all lanes. Biotinylated (surface) protein and total lysate were probed with anti-DAT antibody for detection of the relative expression level of wild-type and mutant DAT protein. Anti–β-actin antibody showed the relative equivalent loading of total protein.
-
Discussion
To date, inherited infantile parkinsonism syndromes are usually associated with reduced CSF dopamine metabolites and enzyme defects in neurotransmitter pathways (6). Here we describe a distinct type of IPD, with characteristic clinical and unique biochemical features, caused by mutations in SLC6A3. This early-onset complex movement disorder can be mistaken for cerebral palsy and is, to our knowledge, the first instance in which patients with parkinsonian features have been linked to an inherited disorder of dopamine transport.
The pre-synaptic DAT, encoded by SLC6A3, mediates the active reuptake of extraneuronal dopamine and is a principal regulator of the amplitude and duration of dopaminergic action at presynaptic and postsynaptic receptors. DAT is exclusively expressed in dopamine neurons with significantly higher levels of DAT expression in cells of the substantia nigra pars compacta, substantia nigra pars lateralis, and ventral tegmental area (7). Dopamine is transported inward against its concentration gradient using the driving force of the sodium gradient across the plasma membrane. The transport process of dopamine uptake involves translocation of dopamine as well as 2 sodium ions and 1 chloride ion across the cell membrane (8). The Na+/K+ pump thus has a crucial role in generating the electrochemical gradient across the cell membrane and inherited mutations in the gene for the Na+/K+-ATPase α3 subunit (ATP1A3) are associated with autosomal dominant rapid-onset parkinsonism-dystonia (ROPD) (9). Loss-of-function ATP1A3 mutations might thus also possibly indirectly affect DAT function.
Based on the crystal structure of the bacterial transporter analog LeuT (10), the primary substrate site and the sodium binding sites in DAT can be modeled deep in the protein interior, where unwinding of the transmembrane helices 1 and 6 allows extensive interactions among protein residues in transmembrane domains 1, 3, 6, 7, and 8 and substrate or sodium (11). A secondary substrate site has been proposed to be located toward the outside of the transporter (12) in an extracellular vestibule where tricyclic antidepressants also bind in LeuT (2, 13), above the extracellular gate, which, in turn, lies between the 2 substrate sites. Among the positions of the 2 IPD-associated missense mutations reported here, Leu368 in hDAT was in transmembrane domain 7 toward the exterior, many helix turns above the residue (Asn353) that presumably interacts with sodium (Figure 8). Leu368 is 1 helix turn under Met371 and has been shown to be conformationally sensitive to dopamine transport (14). Of the 2 helical portions of extracellular loop 4, EL4b, the position of the second missense mutation, Pro395, is in the helical portion closer to transmembrane 8. Pro395 lies just 1 helix turn above Ala399, a residue that, like Met371, points outward from the entire transporter structure and thus is easily accessible to solvent bathing the outside of the protein (14). The interaction between a sulfhydryl reagent from the outside and position 399 was not blocked in the presence of dopamine, whereas the interaction at the 371 position was highly sensitive to dopamine, which caused conformational changes, burying the residue away from solvent exposure. In consonance, the missense mutation of Pro395 (above 399) had no impact on dopamine binding affinity, whereas the missense mutation of Leu368 (under 371) appeared to interfere with conformational changes occurring with dopamine binding, such that dopamine’s apparent affinity was strongly reduced. Hence, despite the different impact of the 2 missense mutations on dopamine binding affinity, the location of the mutations and the fact that the mutations completely impaired transport of dopamine suggested that both mutations might interfere with conformational changes required during transport. However, a major contributor to reducing dopamine transport appears to be the marked reduction in mature (glycosylated) 85-kDa DAT expression. It has previously been demonstrated that immature non-glycosylated (55-kDa) DAT does not transport dopamine as efficiently as mature 85-kDa DAT (15). Hence a reduction in mature DAT would undoubtedly impair dopamine transport. It is known that glycosylation patterns vary with the cellular environment, which is dependent upon cell type and physiological state of the cell in which the glycoprotein is made, and so analysis of brain DAT glycosylation in DAT IPD cases is of considerable interest.
 Figure 8
Figure 8Two-dimensional representation of DAT topology based on LeuT structure. Twelve transmembrane domains are shown with helically unwound regions in the first and sixth domain; extracellular and intracellular loops include helical portions (e2, e3, e4a, e4b, and i1, i5, respectively) (10, 11). Leu368 (L; red), subject to missense mutation, is shown at the top of transmembrane domain 7, one helix turn under Met371 (M; blue). Pro395 (P; red), also subject to missense mutation, is shown in the e4b portion of extracellular loop 4, one helix turn above Ala399 (A; blue). In the 3-dimensional structure, sodium ions interact not only with residues of transmembrane domains 1, 6, and 8, but also with transmembrane domain 7 (10).
Dopamine mediates a wide array of physiological functions, including locomotion, cognitive processes, neuroendocrine secretion, and the control of motivated behaviors including emotion, affect, and reward mechanisms. DAT is the site of action of drugs such as cocaine and methylphenidate (which inhibit DAT-mediated dopamine reuptake) and amphetamines (16). Dysfunctional dopamine homeostasis has been implicated in a wide number of neurological and neuropsychiatric conditions (17, 18), and the observation that DAT homozygous-null mice exhibit a hyperdopaminergic phenotype with prominent hyperactivity (19) has prompted extensive genetic investigations of SLC6A3 in human neuropsychiatric disorders. Mazei-Robison et al. reported a rare coding variant (A559V) in 2 male siblings with attention deficit hyperactivity disorder (20). The A559V mutant demonstrated increased dopamine efflux (which characteristically is associated with amphetamine-like psychostimulants), and so different genetic alterations in SLC6A3 may affect different facets of DAT function and cause distinct phenotypes. Although the phenotypic features seen in IPD are not evident in the established knockout mouse model, a more highly developed extrapyramidal system in primates may result in differences in attributes of DAT dysfunction in mice and humans.
Although IPD is a rare condition, it is likely underdiagnosed and may mimic mixed forms of cerebral palsy, in which pyramidal features and dystonia coexist. CSF dopamine metabolites are abnormal in IPD and might be used to identify patients with variant phenotypes for SLC6A3 mutation analysis. As the spectrum of SLC6A3 mutations expand, specific genotype-phenotype correlations may emerge. In addition, genetic variants in SLC6A3 may influence susceptibility to other movement disorders such as late-onset parkinsonism and dystonic side effects of anti-parkinsonism drugs or neuroleptics.
Finally, molecular characterization of SLC6A3-associated IPD may provide a basis for the development of effective therapies. Current therapies do not biochemically normalize HVA levels or clinically improve patient symptomatology. L-dopa therapy may be beneficial in other infantile parkinsonism syndromes (such as tyrosine hydroxylase deficiency) with reduced dopamine production. In IPD patients with SLC6A3 mutations, defective DAT function and loss of dopamine reuptake may lead to depletion of neuronal dopamine stores. In fact, data from mice with targeted genetic deletion of dopamine, serotonin, and norepinephrine transporters provide further evidence that these monoamine transporters have an important role in monoamine homeostasis. Such mice show reduced extracellular monoamine clearance rate, decreased amplitude of stimulated monoamine release, elevated basal extracellular monoamine levels, decreased monoamine tissue storage, increased monoamine synthesis rate, and disrupted autoreceptor function (21). Additionally, the reduced reuptake of dopamine by DAT increases the dopamine in the extraneuronal space (as shown by increased HVA in the CSF), which might result in secondary downregulation of postsynaptic dopamine receptor expression. Certainly in the aforementioned monoamine transporter knockout mouse models, there is evidence of postsynaptic receptor downregulation (21). Our patients with DAT IPD demonstrated a poor response to L-dopa, suggesting that therapeutic attempts to normalize brain dopamine levels in DAT IPD might be insufficient to improve function, perhaps because of the complex role of DAT in the fine regulation of amplitude and duration of dopaminergic transmission. Although short-term DAT inhibition appears to potentiate L-dopa therapy in Parkinson disease (22), our experience with DAT IPD suggests that this might not be maintained in the long term. The moderate therapeutic success of DBS in patient 3 may be attributable to the hypothesized role of such targeted neuronal high-frequency stimulation in the modulation of dopamine, glutamate, and GABA neurotransmitter systems within the basal ganglia (23). Such selective neuronal activation and suppression as well as changes in network synchrony may thus result in alleviation of parkinsonian-dystonic symptoms. In the future, trials of DBS as well as partial dopamine receptor agonists/antagonists could be useful for medically ameliorating DAT-associated movement disorders.
-
Methods
Clinical phenotyping
Family members and control subjects provided written informed consent under a research protocol approved by the South Birmingham Local Research Ethics Committee. All 3 affected children with IPD were clinically assessed by a pediatric neurologist. Formal neurological examination of all the parents and unaffected siblings was also undertaken.
Molecular genetic studies
DNA extraction. DNA was extracted from peripheral lymphocytes using standard techniques.
Genome-wide scan. In order to identify common regions of shared homozygosity in affected individuals, a genome-wide scan using the Affymetrix 250K SNP microarray was initially undertaken. DNA was amplified and hybridized to the Affymetrix 250K SNP chips according to the manufacturer’s instructions.
Linkage analysis. All significant regions of common homozygosity identified on genome-wide scan were further investigated using standard polymorphic microsatellite markers (UniSTS; http://www.ncbi.nlm.nih.gov/sites/entrez?db=unists). After PCR amplification of genomic DNA samples, amplified fragments were genotyped using an ABI PRISM 3730 Genetic Analyzer and analyzed using GeneMapper software. Scored genotypes were assembled as haplotypes and analyzed for evidence of linkage.
Gene sequencing. Mutation analysis of SLC6A3 (DAT1) was undertaken by direct sequencing. Primer pairs were designed for exon-specific PCR amplification of the 14 translated exons of the SLC6A3 gene (Table 3). At the purification stage, amplified products were either treated with Exosap or gel-extracted (Qiagen). Sequencing reactions were performed using BigDye Terminator v3.1 Cycle Sequencing kits and run on an ABI PRISM 3730 DNA Analyzer (Applied Biosystems). DNA sequences were viewed and analyzed using Chromas software.
Functional analysis of mutant DAT proteins
Mutant constructs of L368Q hDAT and P395L hDAT were prepared from wild-type pCIN4-hDAT (a gift from Jonathan A. Javitch, Columbia University, New York, New York, USA) as the template for PCR using the QuikChange site-directed mutagenesis kit (Stratagene) with some modifications. Briefly, 50 ng of DNA template (pCIN4-hDAT) was mixed with a primer and its complementary primer (100 ng each), 1 μl dNTP mix, 2.5 μl of 10× reaction buffer, and 1.25 U of PfuTurbo DNA polymerase in a final volume of 25 μl. The PCR conditions included an initial denaturation cycle (1 min, 95°C); 18 cycles of denaturation (30 s, 95°C), annealing (1 min, 55°C), and extension (16 min, 68°C); a final extension cycle (10 min, 68°C); and termination at 4°C. PCR products were digested with 10 U of DpnI at 37°C for 1 h to remove the parental strands. The digested DNA mixture was transformed into E. coli XL-1 blue supercompetent cells by heat shock. Mutagenesis was verified by DNA sequence analysis (by the DNA sequencing facility at the Skirball Institute of Biomolecular Medicine, New York University). The following primers and their complementary primers (IDT Inc.) were used: L368Q, 5′-GTCGTGTTCAGCTTTCAGGGATACATGGCCCAG-3′; P395L, 5′-GATCTTCATTATCTACCTTGAGGCCATCGCTACCC-3′.
Culturing and transient transfection of HEK293 cells with Lipofectamine 2000, was carried out as described previously (2). Uptake of [3H]dopamine into hDAT-expressing cells was measured for 5 minutes at 21°C, as in our previous study (2), with high sodium, low potassium, glucose and tropolone containing buffer (3). Nonspecific uptake was defined by 1 μM CFT. For measurement of cocaine analog binding, cells were incubated with 4 nM [3H]CFT (PerkinElmer) for 20 minutes at 21°C in 200 μl of the same buffer used for the uptake assay. For saturation analysis, increasing concentrations of nonradioactive CFT (0.1–100 nM) were included in assay. The procedures were described previously (4). Nonspecific binding was defined with 1 μM CFT. The Km and Vmax of dopamine uptake and the Kd and maximal binding (Bmax) of [3H]CFT binding were estimated with nonlinear regression fitting using Radlig software (KELL program; Biosoft). The equilibrium dissociation KI of [3H]CFT binding was computed using the Cheng-Prusoff equation (4) from the concentration inhibiting binding by 50%, as estimated using logistic fitting of data by the ORIGIN software (OriginLab Co.). Comparisons of the 2 mutants with wild-type DAT were performed using 1-way ANOVA followed by the Dunnett multiple comparisons test. Statistical significance was considered at P < 0.05.
DAT surface protein preparation by biotinylation and Western blotting
Cells transiently expressing wild-type hDAT, L368Q hDAT, and P395L hDAT and mock cells were processed as described previously (24), with minor changes. Briefly, the cells were washed with cold PBS and incubated with sulfo-NHS-SS-biotin (1 mg/ml PBS; Pierce Biotechnology) for 60 minutes at 4°C, followed by incubation with 50 mM glycine in PBS for 20 minutes and extensive washing. The washed cells were lysed in mammalian protein extraction reagents supplemented with a protein inhibitor cocktail for 10 minutes at 21°C followed by 60 minutes at 4°C with vortexing at 5-minute intervals. The lysate samples were centrifuged at 14,000 g for 15 minutes to remove cell debris. The biotinylated proteins were separated with immobilized monomeric NeutrAvidin (Pierce Biotechnology) and eluted with SDS-PAGE sample buffer. The total lysates and biotinylated proteins were resolved on 4%–12% Bis-tris NuPAGE gels (Invitrogen). DAT was probed with polyclonal anti-DAT antibody (0.3 μg/ml) against the C-terminal of DAT (Millipore), followed by HRP-conjugated goat anti-rabbit antibody. Polyclonal anti–β-actin antibody (Sigma-Aldrich) was used as an internal control for loading. The transporter signal was visualized using SuperSignal West Dura Extended Duration chemiluminescence substrate solution (Thermo Scientific).
-
Acknowledgments
We thank the Birmingham Children’s Hospital Research Foundation, NewLife, Action Medical Research, the Wellcome Trust, WellChild, and the NIH for financial support. We also thank Robert Surtees.
Address correspondence to: Eamonn R. Maher, Department of Medical and Molecular Genetics, Institute of Biomedical Research, Birmingham University, Edgbaston, Birmingham B15 2TT, United Kingdom. Phone: 00-44-121-627-2741; Fax: 00-44-121-414-2538; E-mail: e.r.maher@bham.ac.uk.
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: CFT, 2β-carbomethoxy-3β-(4-fluorophenyl)-tropane; CSF, cerebrospinal fluid; DAT, dopamine transporter; DBS, deep brain stimulator; hDAT, human DAT; HVA, homovanillic acid; IPD, infantile parkinsonism-dystonia.
Reference information: J. Clin. Invest.119:1595–1603 (2009). doi:10.1172/JCI39060
See the related article at Infantile parkinsonism-dystonia: a dopamine “transportopathy”.
-
References
- Assmann, B.E., et al. 2004. Infantile Parkinsonism-dystonia and elevated dopamine metabolites in CSF. Neurology. 62:1872-1874. View this article via: PubMed Google Scholar
- Zhou, Z., et al. 2007. LeuT-desipramine structure reveals how antidepressants block neurotransmitter reuptake. Science. 317:1390-1393.
- Chen, N., Rickey, J., Berfield, J.L., Reith, M.E.A. 2004. Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation and cocaine binding. J. Biol. Chem. 279:5508-5519.
- Chen, N., Reith, M.E. 2007. Substrates and inhibitors display different sensitivity to expression level of the dopamine transporter in heterologously expressing cells. J. Neurochem. 101:377-388.
- Chen, N., Zhen, J., Reith, M.E. 2004. Mutation of Trp84 and Asp313 of the dopamine transporter reveals similar mode of binding interaction for GBR12909 and benztropine as opposed to cocaine. J. Neurochem. 89:853-864.
- Pearl, P.L., Taylor, J.L., Trzcinski, S., Sokohl, A. 2007. .T he paediatric neurotransmitter disorders. J. Child Neurol. 22:606-616.
- Storch, A., Ludolph, A.C., Schwarz, J. 2004. Dopamine transporter: involvement in selective dopaminergic neurotoxicity and degeneration. J. Neural Transm. 111:1267-1286.
- McElvain, J.S., Schenk, J.O. 1992. A multisubstrate mechanism of striatal dopamine uptake and its inhibition by cocaine. Biochem. Pharmacol. 43:2189-2199.
- de Carvalho Aguiar, P., et al. 2004. Mutations in the Na+/K+ -ATPase alpha3 gene ATP1A3 are associated with rapid-onset dystonia parkinsonism. Neuron. 43:169-175. View this article via: PubMed Google Scholar
- Yamashita, A., Singh, S.K., Kawate, T., Jin, Y., Gouaux, E. 2005. Crystal structure of a bacterial homologue of Na+/Cl––dependent neurotransmitter transporters. Nature. 437:215-223.
- Beuming, T., Shi, L., Javitch, J.A., Weinstein, H. 2006. A comprehensive structure-based alignment of prokaryotic and eukaryotic neurotransmitter/Na+ symporters (NSS) aids in the use of the LeuT structure to probe NSS structure and function. Mol. Pharmacol. 70:1630-1642.
- Shi, L., Quick, M., Zhao, Y., Weinstein, H., Javitch, J.A. 2008. The mechanism of a neurotransmitter:sodium symporter–inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol. Cell. 30:667-677.
- Singh, S.K., Yamashita, A., Gouaux, E. 2007. Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature. 448:952-956.
- Norregaard, L., Loland, C.J., Gether, U. 2005. Evidence for distinct sodium-, dopamine-, and cocaine-dependent conformational changes in transmembrane segments 7 and 8 of the dopamine transporter. J. Biol. Chem. 278:30587-30596.
- Li, L.B., et al. 2004. The role of N-glycosylation in function and surface trafficking of the human dopamine transporter. J. Biol. Chem. 279:21012-21020.
- Bannon, M.J. 2005. The dopamine transporter: role in neurotoxicity and human disease. Toxicol. Appl. Pharmacol. 204:355-360.
- Mehler-Wex, C., Riederer, P., Gerlach, M. 2006. Dopaminergic dysbalance in distinct basal ganglia neurocircuits: implications for the pathophysiology of Parkinson’s disease, schizophrenia and attention deficit hyperactivity disorder. Neurotox. Res. 10:167-179. View this article via: PubMed Google Scholar
- Mazei-Robinson, M.S., Blakely, R.D. 2006. ADHD and the dopamine transporter: are there reasons to pay attention? Handb. Exp. Pharmacol. 175:373-415.
- Giros, B., Jaber, M., Jones, S.R., Wightman, R.M., Caron, M.G. 1996. Hyperlocomotion and indifference to cocaine and amphetamine in mice lacking the dopamine transporter. Nature. 379:606-612.
- Mazei-Robison, M.S., et al. 2008. Anomalous dopamine release associated with a human dopamine transporter coding variant. J. Neurosci. 28:7040-7046.
- Gainetdinov, R.R., Caron, M.G. 2003. Monomaine transporters: from genes to behavior. Annu. Rev. Pharmacol. Toxicol. 43:261-284.
- Nutt, J.G., Carter, J.H., Sexton, G.J. 2004. The dopamine transporter: importance in Parkinson’s disease. Ann. Neurol. 55:766-773.
- McIntyre, C.C., Savasta, M., Walter, B.L., Vitek, J.L. 2004. How does deep brain stimulation work? Present understanding and future questions. J. Clin. Neurophysiol. 21:40-50.
- Chen, N., Reith, M.E. 2008. Substrates dissociate dopamine transporter oligomers. J. Neurochem. 105:910-920. View this article via: PubMed Google Scholar
- Assmann, B.E., et al. 2004. Infantile Parkinsonism-dystonia and elevated dopamine metabolites in CSF. Neurology. 62:1872-1874.
-
Version history
- Version 1 (May 26, 2009): No description
- Version 2 (June 1, 2009): No description



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)











