Advertisement
Commentary Free access | 10.1172/JCI135713
CoRESTed development of regulatory T cells
Luisa Morales-Nebreda,1 Kathryn A. Helmin,1 and Benjamin D. Singer1,2,3
1Division of Pulmonary and Critical Care Medicine,
2Department of Biochemistry and Molecular Genetics, and
3Simpson Querrey Center for Epigenetics, Northwestern University Feinberg School of Medicine, Chicago, Illinois, USA.
Address correspondence to: Benjamin D. Singer, Division of Pulmonary and Critical Care Medicine, Department of Biochemistry and Molecular Genetics, Northwestern University Feinberg School of Medicine, Simpson Querrey 5th Floor, 303 E. Superior Street, Chicago, Illinois, 60611, USA. Phone: 312.503.4494; Email: benjamin-singer@northwestern.edu.
Find articles by
Morales-Nebreda, L.
in:
PubMed
|
Google Scholar
|

1Division of Pulmonary and Critical Care Medicine,
2Department of Biochemistry and Molecular Genetics, and
3Simpson Querrey Center for Epigenetics, Northwestern University Feinberg School of Medicine, Chicago, Illinois, USA.
Address correspondence to: Benjamin D. Singer, Division of Pulmonary and Critical Care Medicine, Department of Biochemistry and Molecular Genetics, Northwestern University Feinberg School of Medicine, Simpson Querrey 5th Floor, 303 E. Superior Street, Chicago, Illinois, 60611, USA. Phone: 312.503.4494; Email: benjamin-singer@northwestern.edu.
Find articles by Helmin, K. in: PubMed | Google Scholar
1Division of Pulmonary and Critical Care Medicine,
2Department of Biochemistry and Molecular Genetics, and
3Simpson Querrey Center for Epigenetics, Northwestern University Feinberg School of Medicine, Chicago, Illinois, USA.
Address correspondence to: Benjamin D. Singer, Division of Pulmonary and Critical Care Medicine, Department of Biochemistry and Molecular Genetics, Northwestern University Feinberg School of Medicine, Simpson Querrey 5th Floor, 303 E. Superior Street, Chicago, Illinois, 60611, USA. Phone: 312.503.4494; Email: benjamin-singer@northwestern.edu.
Find articles by
Singer, B.
in:
PubMed
|
Google Scholar
|

Published March 3, 2020 - More info
J Clin Invest. 2020;130(4):1618–1621. https://doi.org/10.1172/JCI135713.
© 2020 American Society for Clinical Investigation
Related article:
Abstract
Foxp3+ Tregs are key to immune homeostasis, but the contributions of various large, multiprotein complexes that regulate gene expression remain unexplored. We analyzed the role in Tregs of the evolutionarily conserved CoREST complex, consisting of a scaffolding protein, Rcor1 or Rcor2, plus Hdac1 or Hdac2 and Lsd1 enzymes. Rcor1, Rcor2, and Lsd1 were physically associated with Foxp3, and mice with conditional deletion of Rcor1 in Foxp3+ Tregs had decreased proportions of Tregs in peripheral lymphoid tissues and increased Treg expression of IL-2 and IFN-γ compared with what was found in WT cells. Mice with conditional deletion of the gene encoding Rcor1 in their Tregs had reduced suppression of homeostatic proliferation, inability to maintain long-term allograft survival despite costimulation blockade, and enhanced antitumor immunity in syngeneic models. Comparable findings were seen in WT mice treated with CoREST complex bivalent inhibitors, which also altered the phenotype of human Tregs and impaired their suppressive function. Our data point to the potential for therapeutic modulation of Treg functions by pharmacologic targeting of enzymatic components of the CoREST complex and contribute to an understanding of the biochemical and molecular mechanisms by which Foxp3 represses large gene sets and maintains the unique properties of this key immune cell.
Authors
Yan Xiong, Liqing Wang, Eros Di Giorgio, Tatiana Akimova, Ulf H. Beier, Rongxiang Han, Matteo Trevisanut, Jay H. Kalin, Philip A. Cole, Wayne W. Hancock
-
Abstract
Tregs require specific epigenetic signatures to induce and maintain their suppressive function in the context of inflammation and cancer surveillance. In this issue of the JCI, Xiong and colleagues identify a critical role for the epigenetic repressor REST corepressor 1 (CoREST) in promoting Treg suppressive transcriptional and functional programs. Pharmacologic inhibition and genetic loss of CoREST in Tregs impaired organ allograft tolerance and unleashed antitumor immunity via epigenetic activation of effector T cell programs. We propose that exploiting epigenetic control mechanisms will further the translation of Treg-based therapeutics to target inflammatory and malignant disorders.
-
Epigenetic landscape of Treg development and function
Immunomodulatory Tregs, a CD4+ T cell subset, are essential for maintaining organ homeostasis and suppressing immunopathology (1, 2). The FOXP3 transcription factor specifies the identity and suppressive function of Tregs. Notably, mice harboring a mutation in the Foxp3 gene exhibit the scurfy phenotype, succumbing to multiorgan lymphoproliferative inflammation approximately 4 weeks after birth (3). Loss-of-function mutations in the human FOXP3 gene result in the autoimmune syndrome immune dysregulation, polyendocrinopathy, enteropathy X-linked (IPEX) (4). Treg dysfunction also contributes to the pathogenesis of organ allograft rejection and common autoimmune syndromes, including systemic lupus erythematosus and systemic sclerosis (5, 6). Conversely, cancer immunotherapy seeks to inhibit Treg suppressive function to uncheck effector T cell–mediated killing of malignant cells (7).
A permissive epigenetic landscape tightly regulates lymphoid cell specification programs to allow the recruitment of cell type–specific transcription factors. The reversibility and cue-sensing nature of epigenetic phenomena, such as DNA methylation and histone modifications, shape chromatin topology to regulate gene expression and allow context-dependent functional adaptability while maintaining lineage stability. Indeed, genome-wide studies have shown differing DNA methylation and histone modification patterns that distinguish Treg from conventional T cell development (8). Along with FOXP3 expression, Tregs independently require a specific DNA hypomethylation pattern at regulatory elements of key genes involved with their suppressive function (8, 9). DNA hypomethylation of enhancers at the Foxp3 locus plays a critical role in supporting Treg differentiation, expansion, and stability. Moreover, the global chromatin organizer Satb1 binds genomic regions known as super-enhancers, which are enriched for active enhancer elements that control Treg signature genes, including Foxp3 (10). Upon Satb1 binding, chromatin loop formation ensues, allowing distal regulatory elements to interact and recruit transcription factors and epigenetic modifiers that activate and stabilize the Treg-specific gene regulatory network. In addition, similarly to conventional CD4+ T cells, Tregs can acquire effector phenotypes (e.g., Th1, Th2, and Th17) in response to environmental cues. Accordingly, both permissive and repressive epigenetic marks in Tregs associate with the expression of effector subset-specific transcription factors, cytokines, and chemokines tailored to mitigate distinct types of inflammation.
Collectively, Tregs require a highly specific epigenome that specifies their identity, yet remains adaptable to Th-polarizing stimuli, in order to promote the myriad functions Tregs are capable of exerting. Therefore, it is not surprising that deregulation of different epigenomic writers, readers, and erasers results in loss of Treg identity and function to cause human disease.
-
Chromatin repressive complexes and Treg biology
In the current issue of the JCI, Xiong and colleagues explored the role of the epigenome-modifying complex REST corepressor 1 (CoREST) in Treg development and function (11). This large multimeric complex serves as a scaffold for the binding of transcription factors and epigenome-modifying enzymes, including histone deacetylases (HDAC1 and HDAC2) that remove acetyl groups from histone tails as well as lysine-specific histone demethylase 1A (LSD1), which catalyzes histone 3 lysine 4 mono- and di-methyl (H3K4me1/2) demethylation. Histone deacetylases have been previously implicated in Treg development and function. In a mouse model, Treg-specific deletion of HDAC3 resulted in impaired Treg suppressive capacity and lethal autoimmunity. Gene expression profiling of these HDAC3-deficient Tregs revealed enrichment of proinflammatory cytokines, chemokines, and adhesion-related receptors (12). Conversely, Treg-specific deletion of HDAC9 or HDAC6 enhances Treg suppressive function (13). These findings demonstrate the complexity of epigenetic networks and underscore the need for detailed data sets describing the function of key epigenetic regulators such as CoREST in a Treg-specific context.
Although the CoREST complex has a well-established role in regulating the hematopoietic system during embryonic development, its role in modulating Treg identity and function remains largely unexplored. To that end, Xiong and colleagues found that under stimulating conditions, Treg-specific deletion of CoREST (also known as Rcor1) led to phenotypic and functional reprogramming, with upregulation of proinflammatory transcription factors, cytokines, and chemokine receptors, such as T-bet, STAT1, IL-2, IFN-γ, and CXCR3. Their findings revealed that loss of CoREST decreased recruitment of repressive histone-modifying enzymes to the promoters of genes encoding these molecules, resulting in increased deposition of permissive histone marks (e.g., H3K9Ac and H3K4me2) and enhanced proinflammatory gene expression. Using adoptive transfer of Rcor1-deficient Tregs, they went on to determine that disruption of the CoREST complex results in substantial loss of Treg-mediated suppression of effector responses in murine models of tumor immunity and cardiac allograft tolerance. Indeed, Rcor1-deficient Tregs promoted antitumor activity and failed to induce long-term survival of cardiac allografts. Moreover, pharmacologic inhibition of the CoREST complex in murine and human Tregs led to reduced expression of suppressive genes, increased expression of Th1 response-associated genes, and diminished suppressive function (11).
FOXP3 forms multimeric complexes with several hundred partners, including CoREST, to induce Treg-specific gene expression programs and repress effector T cell programs. The association with several higher-order structures of chromatin complexes typifies cell lineage–specifying transcription factors such as FOXP3 (14). Thus, pharmacotherapies or epigenome editing technologies could target interactions between complexes such as CoREST or individual components of a specific complex within Tregs to mitigate or promote immune tolerance in malignant or inflammatory contexts, respectively (Figure 1).
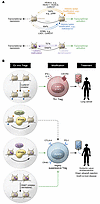 Figure 1
Figure 1Pharmacologic and epigenome editing approaches to Treg immunotherapy. (A) Epigenetic writers and erasers define the epigenetic landscape and control transcription. (B) Ex vivo modification using pharmacologic or epigenome editing to alter the epigenetic landscape could generate Tregs that are fitted to a particular clinical context. Th1 Tregs could be infused to treat malignant disorders, and highly suppressive Tregs could be infused to treat inflammatory disorders such as systemic lupus erythematosus, organ allograft rejection, or graft-versus-host disease. HAT, histone acetyltransferase; TET, ten-eleven translocase (DNA demethylase); KMT, lysine methyltransferase; KDM, lysine demethylase.
-
The next frontier: epigenetic reprogramming of Treg-based therapy
Targeting the epigenetic landscape is a widely recognized strategy for managing malignant disorders in the clinical setting. Pharmacologic reprogrammers of the epigenome include DNA methyltransferase inhibitors (DNMTis), histone deacetylase inhibitors (HDACis), histone methyltransferase inhibitors (e.g., EZH2 inhibitors), and bromodomain and extraterminal protein motifs (iBETs). DNMTis constitute the mainstay therapy for myelodysplastic syndrome and some forms of acute myeloid leukemia (8). HDACis have also been approved for use in several hematologic and solid tumors. In addition to malignancy, these compounds hold promise along with iBETs in the management of acute and chronic inflammatory diseases due to their pleotropic effects within the immune system (13, 15).
Nevertheless, challenges remain with the use of small-molecule epigenome–modifying drugs. For example, available DNMTis do not target a specific family member (i.e., DNMT1, DNMT3A, or DNMT3B), and evidence supports different functional effects of pharmacologic pan-DNMT inhibition versus genetic loss of individual DNMT family members within Tregs (16). Even selective or isoform-specific HDACis can affect distinct HDAC-containing remodeling complexes. For example, while Treg-specific HDAC1 ablation dampens Treg suppressive function in an allograft tolerance murine model, deletion of HDAC2 enhances Treg suppressive function. Moreover, pharmacologic inhibition of HDAC1-3 failed to modulate Treg function in a murine colitis model (17). Thus, there is substantial variability, unpredictability, and potential for off-target toxicity of these broad epigenome reprogrammers. We suggest that focus should be applied to more specific epigenetic targets, as Xiong and colleagues have shown with CoREST inhibitors (11).
Ex vivo modification of Tregs followed by therapeutic cell transfer represents a strategy for addressing malignant and inflammatory disorders (18). Successful T cell transfer therapy was first described over 40 years ago when autologous T cells were administered to patients with metastatic melanoma (19). Currently, over 50 clinical trials are examining Treg transfer therapy to manage autoimmune diseases and induce tolerance upon hematopoietic stem cell or solid organ transplantation (20). Although results from many of these trials have confirmed the safety and feasibility of Treg transfer, limitations as to the durability, specificity, and phenotypic and functional stability of the administered cells remain a foreseeable challenge (18). Genome-editing technologies, such as the CRISPR-Cas9 system, might be used to alter locus-specific epigenetic signatures, harnessing unprecedented potential as a therapeutic strategy. Investigators have used this editing tool to deposit, recruit, or remove epigenetic marks in T cell populations. For example, dCas9 fused with the catalytic domains of epigenetic writers or erasers can modulate the transcriptional activity of specific loci (21). In addition to manipulating methyl-DNA marks, investigators have introduced dCas9-p300 (dCas9 fused to the histone acetyltransferase p300) and shown increased expression of Treg-specific core signature genes and enhanced suppressive capacity (22). Epigenetic editing tools (e.g., FIRE-Cas9) allow for fast, reversible, and highly locus-specific recruitment of distinct chromatin remodeling complexes, enabling a better understanding of the link between epigenetic modifications and gene expression in disease contexts that could inform the development of unique Treg-based therapies (21).
We envision that the next frontier in Treg-based immunotherapy will involve pharmacologic epigenetic reprogramming or epigenetic editing of specific loci to create Treg transcriptional and functional programs fitted to a clinical context. CRISPR-mediated epigenetic reprogramming of Treg-associated suppressive molecules and expansion of Th1 Tregs could be leveraged to treat malignant diseases. Conversely, manipulation of epigenetic regulatory networks controlling the expression of suppressive Treg effector molecules such as CTLA-4, PD-1, and OX40 could permit a more robust suppressive response during autoimmune diseases, organ allograft rejection, and graft-versus-host disease (Figure 1). Going forward, we believe that a detailed understanding of the molecular networks that underlie and stabilize Treg function will further the translation of epigenome-targeted therapies to the bedside.
-
Acknowledgments
LMN was supported by NIH award T32HL076139. BDS was supported by NIH awards K08HL128867 and U19AI135964.
Address correspondence to: Benjamin D. Singer, Division of Pulmonary and Critical Care Medicine, Department of Biochemistry and Molecular Genetics, Northwestern University Feinberg School of Medicine, Simpson Querrey 5th Floor, 303 E. Superior Street, Chicago, Illinois, 60611, USA. Phone: 312.503.4494; Email: benjamin-singer@northwestern.edu.
-
Footnotes
Conflict of interest: BDS has a pending patent application: US patent 15/542,380, Compositions and Methods to Accelerate Resolution of Acute Lung Inflammation.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(4):1618–1621. https://doi.org/10.1172/JCI135713.
See the related article at Inhibiting the coregulator CoREST impairs Foxp3+ Treg function and promotes antitumor immunity.
-
References
- Sakaguchi S, Mikami N, Wing JB, Tanaka A, Ichiyama K, Ohkura N. Regulatory T cells and human disease. Annu Rev Immunol [published online February 4, 2020]. https://doi.org/10.1146/annurev-immunol-042718-041717.
- Singer BD, Chandel NS. Immunometabolism of pro-repair cells. J Clin Invest. 2019;129(7):2597–2607.
- Brunkow ME, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27(1):68–73.
- Wildin RS, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27(1):18–20.
- Valencia X, Yarboro C, Illei G, Lipsky PE. Deficient CD4+CD25high T regulatory cell function in patients with active systemic lupus erythematosus. J Immunol. 2007;178(4):2579–2588.
- Antiga E, et al. Regulatory T cells in the skin lesions and blood of patients with systemic sclerosis and morphoea. Br J Dermatol. 2010;162(5):1056–1063.
- Simpson TR, et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J Exp Med. 2013;210(9):1695–1710.
- Morales-Nebreda L, McLafferty FS, Singer BD. DNA methylation as a transcriptional regulator of the immune system. Transl Res. 2019;204:1–18.
- Ohkura N, et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37(5):785–799.
- Kitagawa Y, et al. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat Immunol. 2017;18(2):173–183.
- Xiong Y, et al. Inhibiting the coregulator CoREST impairs Foxp3+ Treg function and promotes antitumor immunity. J Clin Invest. 2020;130(4):1830–1842.
- Wang L, et al. FOXP3+ regulatory T cell development and function require histone/protein deacetylase 3. J Clin Invest. 2015;125(3):1111–1123.
- Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov. 2014;13(9):673–691.
- Rudra D, et al. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat Immunol. 2012;13(10):1010–1019.
- McGrath-Morrow SA, et al. DNA methylation regulates the neonatal CD4+ T-cell response to pneumonia in mice. J Biol Chem. 2018;293(30):11772–11783.
- Wang L, et al. Foxp3+ T-regulatory cells require DNA methyltransferase 1 expression to prevent development of lethal autoimmunity. Blood. 2013;121(18):3631–3639.
- Wang L, et al. Histone/protein deacetylase inhibitor therapy for enhancement of Foxp3+ T-regulatory cell function posttransplantation. Am J Transplant. 2018;18(7):1596–1603.
- Singer BD, King LS, D’Alessio FR. Regulatory T cells as immunotherapy. Front Immunol. 2014;5:46. View this article via: PubMed Google Scholar
- Rosenberg SA, et al. Use of tumor-infiltrating lymphocytes and interleukin-2 in the immunotherapy of patients with metastatic melanoma. A preliminary report. N Engl J Med. 1988;319(25):1676–1680.
- Ferreira LMR, Muller YD, Bluestone JA, Tang Q. Next-generation regulatory T cell therapy. Nat Rev Drug Discov. 2019;18(10):749–769.
- Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9(1):1911.
- Okada M, Kanamori M, Someya K, Nakatsukasa H, Yoshimura A. Stabilization of Foxp3 expression by CRISPR-dCas9-based epigenome editing in mouse primary T cells. Epigenetics Chromatin. 2017;10:24. View this article via: PubMed Google Scholar
-
Version history
- Version 1 (March 3, 2020): Electronic publication
- Version 2 (April 1, 2020): Print issue publication



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)

