Advertisement
Research ArticleMetabolismOncology Free access | 10.1172/JCI128287
mTORC1 feedback to AKT modulates lysosomal biogenesis through MiT/TFE regulation
Kaushal Asrani,1 Sanjana Murali,1 Brandon Lam,1 Chan-Hyun Na,2 Pornima Phatak,3 Akshay Sood,1 Harsimar Kaur,1 Zoya Khan,1 Michaël Noë,1 Ravi K. Anchoori,1 C. Conover Talbot Jr.,4 Barbara Smith,5 Michael Skaro,1 and Tamara L. Lotan1
1Department of Pathology and
2Institute for Cell Engineering, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Baltimore Veteran Affairs Medical Center, Baltimore, Maryland, USA.
4Institute for Basic Biomedical Sciences and
5Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Tamara Lotan and Kaushal Asrani, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.614.9196; Email: tlotan1@jhmi.edu, kasrani1@jhmi.edu.
Find articles by Asrani, K. in: PubMed | Google Scholar
1Department of Pathology and
2Institute for Cell Engineering, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Baltimore Veteran Affairs Medical Center, Baltimore, Maryland, USA.
4Institute for Basic Biomedical Sciences and
5Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Tamara Lotan and Kaushal Asrani, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.614.9196; Email: tlotan1@jhmi.edu, kasrani1@jhmi.edu.
Find articles by Murali, S. in: PubMed | Google Scholar
1Department of Pathology and
2Institute for Cell Engineering, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Baltimore Veteran Affairs Medical Center, Baltimore, Maryland, USA.
4Institute for Basic Biomedical Sciences and
5Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Tamara Lotan and Kaushal Asrani, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.614.9196; Email: tlotan1@jhmi.edu, kasrani1@jhmi.edu.
Find articles by Lam, B. in: PubMed | Google Scholar
1Department of Pathology and
2Institute for Cell Engineering, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Baltimore Veteran Affairs Medical Center, Baltimore, Maryland, USA.
4Institute for Basic Biomedical Sciences and
5Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Tamara Lotan and Kaushal Asrani, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.614.9196; Email: tlotan1@jhmi.edu, kasrani1@jhmi.edu.
Find articles by Na, C. in: PubMed | Google Scholar
1Department of Pathology and
2Institute for Cell Engineering, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Baltimore Veteran Affairs Medical Center, Baltimore, Maryland, USA.
4Institute for Basic Biomedical Sciences and
5Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Tamara Lotan and Kaushal Asrani, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.614.9196; Email: tlotan1@jhmi.edu, kasrani1@jhmi.edu.
Find articles by Phatak, P. in: PubMed | Google Scholar
1Department of Pathology and
2Institute for Cell Engineering, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Baltimore Veteran Affairs Medical Center, Baltimore, Maryland, USA.
4Institute for Basic Biomedical Sciences and
5Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Tamara Lotan and Kaushal Asrani, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.614.9196; Email: tlotan1@jhmi.edu, kasrani1@jhmi.edu.
Find articles by
Sood, A.
in:
PubMed
|
Google Scholar
|

1Department of Pathology and
2Institute for Cell Engineering, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Baltimore Veteran Affairs Medical Center, Baltimore, Maryland, USA.
4Institute for Basic Biomedical Sciences and
5Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Tamara Lotan and Kaushal Asrani, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.614.9196; Email: tlotan1@jhmi.edu, kasrani1@jhmi.edu.
Find articles by Kaur, H. in: PubMed | Google Scholar
1Department of Pathology and
2Institute for Cell Engineering, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Baltimore Veteran Affairs Medical Center, Baltimore, Maryland, USA.
4Institute for Basic Biomedical Sciences and
5Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Tamara Lotan and Kaushal Asrani, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.614.9196; Email: tlotan1@jhmi.edu, kasrani1@jhmi.edu.
Find articles by Khan, Z. in: PubMed | Google Scholar
1Department of Pathology and
2Institute for Cell Engineering, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Baltimore Veteran Affairs Medical Center, Baltimore, Maryland, USA.
4Institute for Basic Biomedical Sciences and
5Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Tamara Lotan and Kaushal Asrani, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.614.9196; Email: tlotan1@jhmi.edu, kasrani1@jhmi.edu.
Find articles by
Noë, M.
in:
PubMed
|
Google Scholar
|

1Department of Pathology and
2Institute for Cell Engineering, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Baltimore Veteran Affairs Medical Center, Baltimore, Maryland, USA.
4Institute for Basic Biomedical Sciences and
5Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Tamara Lotan and Kaushal Asrani, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.614.9196; Email: tlotan1@jhmi.edu, kasrani1@jhmi.edu.
Find articles by Anchoori, R. in: PubMed | Google Scholar
1Department of Pathology and
2Institute for Cell Engineering, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Baltimore Veteran Affairs Medical Center, Baltimore, Maryland, USA.
4Institute for Basic Biomedical Sciences and
5Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Tamara Lotan and Kaushal Asrani, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.614.9196; Email: tlotan1@jhmi.edu, kasrani1@jhmi.edu.
Find articles by
Talbot, C.
in:
PubMed
|
Google Scholar
|

1Department of Pathology and
2Institute for Cell Engineering, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Baltimore Veteran Affairs Medical Center, Baltimore, Maryland, USA.
4Institute for Basic Biomedical Sciences and
5Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Tamara Lotan and Kaushal Asrani, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.614.9196; Email: tlotan1@jhmi.edu, kasrani1@jhmi.edu.
Find articles by Smith, B. in: PubMed | Google Scholar
1Department of Pathology and
2Institute for Cell Engineering, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Baltimore Veteran Affairs Medical Center, Baltimore, Maryland, USA.
4Institute for Basic Biomedical Sciences and
5Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Tamara Lotan and Kaushal Asrani, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.614.9196; Email: tlotan1@jhmi.edu, kasrani1@jhmi.edu.
Find articles by Skaro, M. in: PubMed | Google Scholar
1Department of Pathology and
2Institute for Cell Engineering, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Baltimore Veteran Affairs Medical Center, Baltimore, Maryland, USA.
4Institute for Basic Biomedical Sciences and
5Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Tamara Lotan and Kaushal Asrani, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.614.9196; Email: tlotan1@jhmi.edu, kasrani1@jhmi.edu.
Find articles by Lotan, T. in: PubMed | Google Scholar
Published September 17, 2019 - More info
J Clin Invest. 2019;129(12):5584–5599. https://doi.org/10.1172/JCI128287.
© 2019 American Society for Clinical Investigation
Received: February 20, 2019; Accepted: September 10, 2019
-
Abstract
The microphthalmia family of transcription factors (MiT/TFEs) controls lysosomal biogenesis and is negatively regulated by the nutrient sensor mTORC1. However, the mechanisms by which cells with constitutive mTORC1 signaling maintain lysosomal catabolism remain to be elucidated. Using the murine epidermis as a model system, we found that epidermal Tsc1 deletion resulted in a phenotype characterized by wavy hair and curly whiskers, and was associated with increased EGFR and HER2 degradation. Unexpectedly, constitutive mTORC1 activation with Tsc1 loss increased lysosomal content via upregulated expression and activity of MiT/TFEs, whereas genetic deletion of Rheb or Rptor or prolonged pharmacologic mTORC1 inactivation had the reverse effect. This paradoxical increase in lysosomal biogenesis by mTORC1 was mediated by feedback inhibition of AKT, and a resulting suppression of AKT-induced MiT/TFE downregulation. Thus, inhibiting hyperactive AKT signaling in the context of mTORC1 loss-of-function fully restored MiT/TFE expression and activity. These data suggest that signaling feedback loops work to restrain or maintain cellular lysosomal content during chronically inhibited or constitutively active mTORC1 signaling, respectively, and reveal a mechanism by which mTORC1 regulates upstream receptor tyrosine kinase signaling.
-
Introduction
The microphthalmia family of transcription factors (MiT/TFEs) is composed of 4 conserved members (Mitf/Tfe3/Tfeb/Tfec) that are essential regulators of lysosomal biogenesis and autophagy. MiTs are functionally redundant and regulate transcription at CLEAR (coordinated lysosomal expression and regulation) motifs on lysosomal/autophagy target genes. The regulation of MiT/TFE transcriptional activity is complex and understood to be governed by short-term subcellular localization changes driven principally by mTORC1 kinase signaling (1). According to current models, phosphorylation of MiT/TFE proteins by mTORC1 leads to their cytoplasmic retention, resulting in decreased lysosomal biogenesis (2–6). This is consistent with the known role of mTORC1, a key sensor of cellular nutrient levels, in the negative regulation of autophagy (7).
However, this model of MiT/TFE regulation raises an important question: how can cells maintain lysosomal content in the face of persistent mTORC1 signaling? Upregulated mTORC1 activity and lysosomal biogenesis must coexist during physiological states such as recovery from starvation (8) and physical exercise (9, 10). Strikingly, several lines of evidence suggest that constitutive/prolonged mTORC1 activity may itself paradoxically activate lysosomal biogenesis via increased MiT/TFE activity. In a small number of studies, constitutive mTORC1 hyperactivity (via Tsc1/2 loss) positively regulated transcription factor EB (TFEB)-dependent lysosomal genes (11) and promoted TFE3 nuclear localization in an mTORC1-dependent manner (12, 13), through undefined mechanisms. Furthermore, MiT/TFEs themselves stimulate mTORC1 activity in multiple cell types in response to nutrients, though their effect on cells with constitutive mTORC1 activation is less certain (14). These findings suggest the intriguing possibility of an mTORC1-MiT/TFE–positive feedback loop. Notably, MiT/TFE activity is also coregulated by numerous oncogenic pathways in parallel to mTORC1, including ERK, GSK3, PKC, and AKT (15–17). Taken together, these data raise the likelihood that mTORC1 regulation of MiT/TFE activity is more complex than previously appreciated.
As a first step to understanding how mTORC1 regulates MiT/TFE activity, we studied isogenic normal cells with or without genetic perturbations leading to constitutive or abrogated mTORC1 signaling. The epidermis and primary keratinocyte cultures provide a unique and well-characterized epithelial model system where the lysosome plays an important role in cellular differentiation and homeostasis (18), thus we developed genetically engineered mouse models of Tsc1, Rheb, or Rptor conditional deletion in the epidermis. Herein, we demonstrate that in the context of long-term, bidirectional mTORC1 signaling perturbation, mTORC1 feedback to AKT prevails to regulate MiT/TFE levels and lysosomal biogenesis. These findings begin to explain how constitutive mTORC1 activation may upregulate lysosomal catabolism and provide a mechanism by which mTORC1 signaling feedback modulates upstream EGFR and HER2 activity.
-
Results
Epidermal mTORC1 gain-of-function models have skin defects reminiscent of epidermal EGFR or TGF-α loss. Germline inactivation of Tsc1 is associated with embryonic lethality (19). To study mTORC1 function in the epidermis, we examined mice with conditional deletion of epidermal Tsc1 by crossing floxed Tsc1 mice (Tsc1fl/fl) with Krt14-Cre mice (which express Cre recombinase driven by the keratin 14 promoter in the basal epidermis by E14.5), to generate Tsc1fl/fl/Krt14-Cre mice (Tsc1-cKO). The presence of Tsc1fl/fl alleles and Krt14-Cre was confirmed by PCR genotyping (Figure 1A). TSC1 loss was verified by immunoblots from epidermal lysates (Figure 1B). In addition, we also prepared parallel primary keratinocyte cultures from these mice to further allow in vitro perturbation experiments in this system and confirm all in vivo findings (Figure 1B). Tsc1-cKO mice were viable and born in the expected Mendelian ratios. However, they could be distinguished by curly vibrissae at birth and coarse, wavy fur by 4 weeks (Figure 1C). During this period, Tsc1-cKO mice developed epidermal thickening and showed increased p-S6 levels by immunofluorescence, consistent with increased mTORC1 activity (Figure 1D). By 6 months, Tsc1-cKO mice had hair loss and severe facial inflammation (Figure 1E), a phenotype strikingly similar to murine epidermal TGF-α or EGFR loss (20–22). To verify mTORC1 dependency of this phenotype, we crossed K14-cre mice with Rheb S16Hfl/fl mice, which express a constitutively active Rheb transgene resistant to TSC GTPase-activating protein (GAP) activity expressed upon Cre excision of a loxp-stop-loxp (23). Genotyping PCR confirmed the presence of S16Hfl/fl alleles, S16H excision alleles, and Krt14-Cre in Rheb S16H transgenic (Tg) mice (Figure 1F). mTORC1 hyperactivity was confirmed by increased p-S6 levels by epidermal immunofluorescence and keratinocyte immunoblotting (Figure 1G and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI128287DS1). These mice also had wavy fur (Figure 1H), confirming that the Tsc1-cKO phenotype was due to increased Rheb/mTORC1 activity.
 Figure 1
Figure 1Epidermal-specific mTORC1 gain-of-function models have skin defects reminiscent of epidermal EGFR or TGF-α loss. (A) Genotyping PCR of genomic tail DNA from WT and Tsc1-cKO mice showing presence of Tsc1fl/fl alleles and Krt14-Cre in Tsc1-cKO mice. (B) Immunoblotting of WT and Tsc1-cKO epidermal and keratinocyte lysates for Tsc1. (C) Tsc1-cKO pups show curly whiskers at birth (top panel) and wavy fur at 4 weeks (middle, bottom panels), indicated by arrows. (D) Tsc1-cKO mice show thickened epidermis on histology (top panel) and increased mTORC1 activity as seen by p-S6 immunofluorescence (bottom panel). Scale bar: 150 μm. (E) Tsc1-cKO mice develop severe dermatitis in the facial region by 6 months. (F) Genotyping PCR of genomic tail DNA from WT and Rheb1 S16H Tg mice showing presence of Rheb1 S16Hfl/fl alleles, Rheb1 S16H excision alleles, and Krt14-Cre in Rheb1 S16H Tg mice. Rheb1 S16H transgenic mice show increased mTORC1 activity as seen by (G) p-S6 immunofluorescence. Scale bar: 150 μm. (H) Rheb1 S16H transgenic mice show presence of wavy fur, similar to Tsc1-cKO mice.
mTORC1 hyperactivation in Tsc1-cKO epidermis and keratinocytes downregulates EGFR and HER2 protein expression and activity. To assess whether the epidermal phenotype in Tsc1-cKO mice was due to a dysfunction in the EGF pathway, we examined expression of EGFR and its principal binding partner HER2. EGFR and HER2 protein expression were significantly decreased in P7 Tsc1-cKO epidermal lysates (Figure 2A) and keratinocytes (Figure 2B) and in Tsc1fl/fl keratinocytes infected with adenovirus expressing cre recombinase (Tsc1-cre), compared with their respective controls, by immunoblotting (Figure 2C). Within the TSC1-TSC2 complex, TSC1 stabilizes TSC2 while TSC2 acts as a GAP for Rheb, and together the complex modulates mTORC1 activity. TSC2 expression in Tsc1-cKO epidermal lysates was decreased (Supplemental Figure 1B) as previously described (24). In addition, there was decreased EGFR and HER2 protein expression in Tsc2fl/fl keratinocytes infected with adenoviral cre (Supplemental Figure 1C). Membrane localized EGFR and HER2 in Tsc1-cKO keratinocytes was also decreased by surface biotinylation assays (Figure 2D). mTORC1 hyperactivity in Tsc1-cKO keratinocytes was confirmed by increased p-S6 levels by immunoblotting, and mTORC1 inhibition using rapamycin or mTOR kinase inhibitors AZD8055 or Torin1 increased EGFR and HER2 protein expression in Tsc1-cKO and Tsc1-cre keratinocytes (Figure 2E and Supplemental Figure 1, D and E). The intensity and duration of EGF-induced EGFR autophosphorylation was diminished in Tsc1-cKO keratinocytes (Figure 2F), with dampened downstream signaling, as shown by decreased basal (Figure 2E, right panel) and EGF-stimulated ERK1/2 and AKT phosphorylation (Figure 2F). Despite these changes in total protein levels, EGFR and HER2 mRNA levels were increased or unchanged in Tsc1-cKO epidermis and keratinocytes (Supplemental Figure 2, A and B), and in Tsc1-cre keratinocytes (Supplemental Figure 2C) compared with their respective controls, suggesting posttranscriptional regulation.
 Figure 2
Figure 2mTORC1 hyperactivation in Tsc1-cKO epidermis and keratinocytes downregulates EGFR and HER2 protein expression and activity. Immunoblotting of (A) WT and Tsc1-cKO epidermal lysates, (B) WT and Tsc1-cKO keratinocyte lysates, and (C) Tsc1fl/fl keratinocyte cultures infected with empty or adenoviral cre recombinase (Tsc1-cre) showing decreased EGFR and HER2 expression with Tsc1 loss (left panels). Immunoblots in B are noncontemporaneous from the same biological replicate, while those in C are contemporaneous and parallel from the same biological replicate. Densitometry quantification of immunoblots (right panels) (biological replicates r ≥ 4; P values are by Student’s t test). Error bars represent SD. (D) Immunoblotting following surface biotinylation and IP showing decreased membrane EGFR and HER2 in Tsc1-cKO keratinocyte lysates compared with WT controls. Na-K ATPase is used to normalize for membrane protein. (E) Immunoblotting of WT and Tsc1-cKO keratinocyte lysates, with or without mTORC1 inhibition using rapamycin (200 nm) or AZD8055 (500 nm), for p-S6, Tsc1, EGFR, and HER2 (left panel) and p-AKT (T308), p-AKT (S473) and p-ERK (right panel). Tsc1-cKO keratinocytes show an increase in p-S6 levels and downregulation of HER2, EGFR, p-AKT, and p-ERK which were rescued upon mTORC1 inhibition. p-S6 and total S6 are noncontemporaneous immunoblots from the same biological replicate. (F) Immunoblotting of serum-starved, EGF-stimulated WT and Tsc1-cKO keratinocyte lysates for EGFR, p-EGFR (Y1068), p-AKT (S473), and p-ERK. The intensity and duration of EGFR autophosphorylation and downstream signaling markers is decreased in Tsc1-cKO keratinocytes.
Genetic and pharmacological inhibition of mTORC1 upregulates EGFR and HER2 protein expression and activity. Studies in cancer cell lines and MEFs have demonstrated that mTORC1 inhibition results in increased PI3K/AKT/MAPK signaling via feedback activation of RTK signaling. This is mediated by mTORC1-dependent phosphorylation of RTK adaptor proteins (IRS-1, GRB10) (25–27) or altered expression of RTKs (IGFR/IR/PDGFR) (28, 29). However, the mechanism of the latter effect on RTK expression is poorly understood. At least one prior study has shown that pharmacologic mTORC1 inhibition also leads to feedback activation of EGFR (30). We examined expression of EGFR and HER2 in mice with conditional epidermal loss of mTORC1 components Rheb or Rptor, as previously described (31). mTORC1 loss-of-function was confirmed by decreased p-p70 S6 kinase and p-4E-BP1 levels in Rptor-cre keratinocyte lysates by immunoblotting (Supplemental Figure 2D). Both Rhebfl/fl/Krt14-Cre (Rheb-cKO) keratinocytes as well as Rptorfl/fl keratinocytes infected with adenoviral cre recombinase (Rptor-cre) upregulated EGFR and HER2 protein expression compared with controls (Figure 3A). Membrane-localized EGFR and HER2 were concomitantly increased by immunofluorescence (Figure 3B) and surface biotinylation assays (Figure 3C). mTORC1 inhibitors (rapamycin, AZD8055, or Torin1) also elevated total (Figure 3D) and membrane (Figure 3E) EGFR and HER2 in WT keratinocytes. mTORC1 inactivation was associated with elevated EGF-stimulated EGFR autophosphorylation in Rheb-cKO (Figure 3F), Rptor-cre (Figure 3G), and AZD8055-treated (Figure 3H and Supplemental Figure 9E) keratinocytes compared with controls, and with elevated and prolonged downstream ERK1/2 and AKT signaling in Rheb-cKO and Rptor-cre keratinocytes (Figure 3F, Supplemental Figure 9A, and ref. 31). Finally, similar to Tsc1-cKO keratinocytes, levels of EGFR and HER2 transcripts were minimally altered in Rptor-cre keratinocytes (Supplemental Figure 2E), indicating a posttranscriptional mechanism of gene regulation. Thus, mTORC1 activity is both necessary and sufficient to regulate EGFR and HER2 total protein expression and activity in the epidermis.
 Figure 3
Figure 3Genetic and pharmacological inhibition of mTORC1 upregulates EGFR and HER2 protein expression and activity. (A) Immunoblotting showing increased expression of EGFR and HER2 in Rheb-cKO (left panel, contemporaneous parallel immunoblots from the same biological replicate) and Rptor-cre (right panel, Rptor and paired actin are noncontemporaneous immunoblots from the same biological replicate) keratinocyte lysates compared with WT/empty controls respectively. (B) Immunofluorescence showing increased membrane EGFR (in Rheb-cKO keratinocytes; left panels) and EGFR and HER2 (in Rptor-cre keratinocytes; right panels) compared with WT/empty controls respectively. Scale bars: 50 μm. (C) Immunoblotting following surface biotinylation and IP showing increased membrane EGFR and HER2 in Rheb-cKO keratinocyte lysates compared with WT controls. Na-K ATPase is used to normalize for membrane protein. (D) Immunoblotting showing increased expression of EGFR and HER2 in rapamycin-treated (R) or AZD8055-treated (A) keratinocyte lysates compared with DMSO-treated (D) controls. p-S6, total S6, p-AKT, and total AKT are noncontemporaneous immunoblots from the same biological replicate. (E) Immunoblotting following surface biotinylation and IP, showing increased membrane EGFR and HER2 in rapamycin-treated (R), AZD8055-treated (A), or Torin1-treated (T) keratinocyte lysates compared with DMSO (D) controls. Enrichment of cell surface proteins in biotin immunoprecipitates is shown using Na-K ATPase. (F) Immunoblotting of serum-starved, EGF-stimulated WT and Rheb-cKO keratinocyte lysates for EGFR, p-EGFR (Y1068), p-AKT (S473), p-AKT (T308), p-ERK, and Rheb. The intensity and duration of EGFR autophosphorylation and downstream signaling markers is increased in Rheb-cKO keratinocytes. EGFR, p-EGFR (Y1068), and Rheb were immunoblotted separately using a different biological replicate. Immunoblotting of serum-starved, EGF-stimulated (G) empty or Rptor-cre and (H) DMSO or AZD8055-treated keratinocyte lysates for EGFR and p-EGFR (Y1068). Empty and Rptor-cre lysates were run on the same gel, separated by a molecular weight marker.
mTORC1 stimulates EGF-induced EGFR degradation by promoting lysosomal biogenesis and activity. EGFR and HER2 levels are downregulated by ligand-induced internalization and lysosomal-mediated degradation (32–34). We analyzed EGFR and HER2 degradation in response to exogenous EGF. Using high-dose EGF to increase the rate of EGF-stimulated EGFR decay in control cells, the EGFR degradation rate was significantly slowed in Rptor-cre keratinocytes (Figure 4, A and B, and Supplemental Figure 3A using low-dose EGF), Rheb-cKO keratinocytes (Supplemental Figure 3, B and C), and AZD8055-treated keratinocytes (Supplemental Figure 3, D and E). Conversely, using low-dose EGF promoted only very minimal EGFR degradation in control keratinocytes, and the rate was significantly enhanced in Tsc1-cKO keratinocytes (Figure 4, C and D). These results raised the possibility that altered lysosomal degradation was mediating the EGFR levels with mTORC1 perturbation. Lysosomes are critical for the degradation of endocytosed or autophagocytosed cellular macromolecules. Lysosomal biogenesis is coordinated by the MiT/TFE subclass of basic helix-loop-helix transcription factors (TFEB/TFE3/MITF/TFEC), which drive transcription from consensus coordinated lysosomal expression and regulation (CLEAR) promoter elements on lysosomal/autophagy genes (5, 15, 35, 36). To investigate lysosomal gene-expression changes downstream of mTORC1 loss-of-function, we performed microarray-based differential expression analysis of E18.5 epidermis from WT/Rptor-cKO mice. Out of 24,697 NCBI gene-annotated coding transcripts, we found 235 genes significantly (greater than 2 SD log2 fold change) upregulated and 941 downregulated in Rptor-cKO compared with Rptor WT epidermis (Supplemental Table 1). We performed GSEA and found that a lysosomal gene signature panel (consisting of 360 lysosomal gene transcripts from the Mouse Lysosome Gene Database [mLGDB; http://lysosome.unipg.it/mouse.php]) was significantly negatively enriched in Rptor-cKO epidermis (Figure 4E). We validated GSEA results by immunoblotting for multiple MiT/TFE-regulated lysosomal proteins, which were downregulated in Rptor-cre and Rheb-cKO keratinocytes compared with their respective controls (Figure 4F and Supplemental Figure 4, A and B). Conversely, lysosomal/autophagy CLEAR target genes (36) were upregulated in Tsc1-cKO keratinocytes by quantitative real-time reverse transcriptase PCR (qRT-PCR) (Figure 5A) and in Tsc1-cKO epidermis (Figure 5B) and keratinocyte (Figure 5C and Supplemental Figure 4C) immunoblots in an mTORC1-dependent manner (Figure 5D). CTSB/LAMP1 immunostaining revealed expansion of both lysosomal organelles in Tsc1-cKO epidermis (Figure 5E). Treatment of Tsc1-cre keratinocytes with the lysosomal V-ATPase inhibitor Bafilomycin A1 was sufficient to rescue EGFR expression, thus suggesting that increased lysosomal expression and/or activity was linked to EGFR loss in cells with constitutive mTORC1 signaling (Figure 5F).
 Figure 4
Figure 4mTORC1 accelerates EGF-induced EGFR degradation. (A) Starved empty and Rptor-cre keratinocytes were stimulated with EGF (50 ng/mL) for the indicated times and immunoblotted for EGFR. (B) EGFR degradation curves. Error bars represent SD. Immunoblots are representative of 3 independent experiments. Empty and Rptor-cre lysates were run on the same gel, separated by a molecular weight marker. (C) Starved WT and Tsc1-cKO keratinocytes were stimulated with EGF (1.5 ng/mL) for the indicated times and immunoblotted for EGFR. (D) EGFR degradation curves. Error bars represent SD. Immunoblots are representative of 3 independent experiments. (E) The Gene Set Enrichment Analysis (GSEA) Enrichment Score Plot depicting the Rptor-cKO versus Rptor WT fold changes of 360 lysosomal genes (from the mouse Lysosome Gene Database [mLGDB]) subset compared with those of all assayed transcripts. The green line is the enrichment score, reflecting the degree of lysosomal genes’ overrepresentation among the Rptor-cKO downregulated (left side) and upregulated (right side) genes. (F) Lysosomal proteins, including those containing a CLEAR-binding motif, are decreased in Rptor-cre keratinocytes compared with empty controls, by immunoblot analyses. Rptor, Ctsd, SQSTM1, Rab7 Laptm4b, and actin (far right panel) are noncontemporaneous immunoblots from the same biological replicate. Densitometry quantification of representative immunoblots from 4 independent experiments are provided in Supplemental Figure 4A.
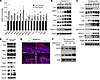 Figure 5
Figure 5mTORC1 activates lysosomal gene expression. (A) qRT-PCR showing upregulation of lysosomal CLEAR target gene transcripts in Tsc1-cre keratinocytes compared with empty controls (r = 4, error bars represent SEM; P values are by Student’s t test). Expression of lysosomal CLEAR gene targets is increased in Tsc1-cKO epidermis (B) and Tsc1-cKO keratinocytes (C), compared with WT controls, by immunoblot analyses. (D) Expression of lysosomal proteins is increased in Tsc1-cKO keratinocytes compared with empty controls, and is downregulated in response to Torin1 (1 μM, 24 hours), by immunoblot analyses. Ctsb, LAMP1, and Ctsd were immunoblotted separately using a different biological replicate. (E) Immunostaining for Ctsb and LAMP1 showing expansion of the lysosomal compartment in basal keratinocytes of Tsc1-cKO epidermis compared with WT controls; white lines demarcate dermal-epidermal junction. Scale bar: 150 μm. (F) Tsc1-cre keratinocytes treated with the lysosomal V-ATPase inhibitor Bafilomycin A1 (100 nm)rescued EGFR expression in a time-dependent manner.
We further characterized lysosomal protein localization by examining expression of lysosomal proteins in lysosomal-enriched fractions of keratinocyte lysates by immunoblotting, and found them to be increased in Tsc1-cre keratinocytes and Rheb1 S16H Tg keratinocytes relative to controls, and decreased in Rptor-cre keratinocytes relative to controls (Figure 6A). Furthermore, the intensity of lysosomal LAMP2 (Figure 6B) and LAMP1 (Figure 6C) immunostaining was decreased in Rptor-cre keratinocytes relative to controls. Quantification of fluorescent intensity demonstrated a significant decrease in mean LAMP1 fluorescence in Rptor-cre keratinocytes (Figure 6D). Finally, to assess the activity of lysosomal enzymes, we incubated cells with Magic Red CTSB, a cathepsin B substrate that produces a cresyl violet fluorophore upon proteolytic cleavage, and measured fluorescence intensity by fluorometry. CTSB activity was significantly decreased in Rptor-cre keratinocytes, relative to controls (Figure 6E).
 Figure 6
Figure 6mTORC1 promotes lysosomal biogenesis and activity. (A) Immunoblot analyses of lysosomal proteins in lysosomal fractions of cellular lysates, showing increased expression of lysosomal proteins in Tsc1-cre keratinocytes (left panel) and Rheb1 S16H Tg keratinocytes (middle panel) compared with controls, and decreased expression of lysosomal proteins in Rptor-cre keratinocytes (right panel) compared with controls. Lysosomal marker Rragc was unaltered across genotypes and used as a loading control. (B) Confocal microscopy analyses of LAMP2 immunostaining demonstrates decreased presence of LAMP2 in Rptor-cre keratinocytes compared with empty controls. Scale bar: 100 μm. (C) Confocal microscopy analyses and double immunostaining for LAMP1/Rptor demonstrates decreased presence of Lamp1 and Rptor in Rptor-cre keratinocytes compared with empty controls. Scale bar: 100 μm. (D) Quantification of LAMP1 fluorescence intensity showing a decrease in mean LAMP1 fluorescence in Rptor-cre keratinocytes compared with controls. The area of LAMP1 was measured using Image J and normalized to the number of nuclei (r = 3, n > 1000). Error bars represent SD, P = 0.0003 by Student’s t test. (E) Lysosomal activity, as measured by fluorometric analyses of cathepsin B activity using the Magic Red Cathepsin B kit, is decreased in Rptor-cre keratinocytes compared with controls (r = 4, error bars represent SD; P = 0.007 by Student’s t test).
mTORC1 drives total MiT/TFE expression, nuclear localization, and CLEAR promoter activity. To understand the basis for altered lysosomal gene expression/activity, we first queried total levels of MiT/TFE proteins, which were increased in Tsc1-cKO epidermis (Figure 7A) and keratinocytes in an mTORC1-sensitive manner (Figure 7B), and correspondingly decreased in Rptor-cKO epidermis (Figure 7C), Rptor-cre keratinocytes (Supplemental Figure 5A), and Rheb-cKO keratinocytes (Supplemental Figure 5B). Furthermore, MiT/TFE proteins were highly enriched in nuclear fraction immunoblots of Tsc1-cKO (Supplemental Figure 5C) and Tsc1-cre (Figure 7, D and E) keratinocytes in an mTORC1-dependent manner. TFE3 was also enriched in the basal nuclei of P7 Tsc1-cKO epidermis by immunohistochemistry (Supplemental Figure 5D), and in the nuclei of Tsc1-cre keratinocytes by immunofluorescence (Figure 7, F and G) in an mTORC1-dependent manner (Supplemental Figure 5E). MiT/TFE proteins were correspondingly decreased in Rptor-cre nuclei by nuclear lysate immunoblots (Supplemental Figure 5, F–H) and immunofluorescence (Supplemental Figure 5I). Importantly, short-term Torin1 treatment (1 hour) promoted, whereas long-term treatment (>24 hours) decreased, nuclear TFE3 by immunofluorescence (Supplemental Figure 6A). We then determined corresponding changes in CLEAR promoter element activity by transfecting cells with a 4X-CLEAR luciferase reporter construct (containing 4 tandem copies of a CLEAR promoter element) (37) and measuring luciferase activity. 4X-CLEAR transactivation was significantly higher in Tsc1-cKO and Tsc1-cre keratinocytes (Figure 7, H and I) and lower in Rptor-cre keratinocytes (Supplemental Figure 6B) compared with their respective controls, linking MiT/TFE levels, CLEAR promoter activity, and altered lysosomal gene expression to mTORC1 status. Finally, in Tsc1-cre keratinocytes, combined MiT/TFE (TFEB/TFE3/MITF) siRNA treatment or single siRNA against these genes repressed expression of many lysosomal CLEAR target genes and proteins, with TFE3 and TFEB appearing to drive most of the effects in triple knockdown treatment (Supplemental Figure 6C, Supplemental Figure 7, and Figure 7J). Combined MiT/TFE knockdown was sufficient to rescue EGFR and HER2 expression (Figure 7J), thus linking increased MiT/TFE transcriptional activity to EGFR and HER2 loss in cells with constitutive mTORC1 signaling.
 Figure 7
Figure 7mTORC1 drives MiT/TFE expression, nuclear localization, and CLEAR promoter activity. (A) Immunoblotting showing increased expression of MiT/TFE proteins in Tsc1-cKO epidermal lysates, and representing the same experiment depicted in Figure 5B. (B) Immunoblotting showing increased expression of MiT/TFE proteins in Tsc1-cre keratinocytes compared with controls, and decreased expression in response to Torin1 (1 μM, 24 hours). TFEB and paired c-Met as well as MITF and paired actin represent contemporaneous parallel immunoblots from the same biological replicate. TFE3 and paired actin were immunoblotted separately using a different biological replicate. (C) Immunoblotting showing decreased expression of MiT/TFE proteins in Rptor-cKO epidermal lysates compared with controls. TFE3 was immunoblotted separately using different biological replicates. (D) MiT/TFE proteins are increased in nuclear-fraction immunoblots of Tsc1-cre keratinocytes compared with controls, and downregulation in response to Torin1 (1 μM, 24 hours). Lamin A/C is used to normalize for nuclear protein. These are contemporaneous parallel immunoblots from the same biological replicate. (E) Densitometry quantification of representative immunoblot experiments shown in D (r ≥ 2; error bars represent SD; P values by 1-way ANOVA). (F) Immunofluorescence showing increased nuclear localization of TFE3 in Tsc1-cre keratinocytes, compared with controls. Scale bar: 150 μm. (G) Quantification of nuclear TFE3 fluorescence from experiments in F (r = 4; n > 1293; P = 0.001 by Student’s t test). 4X-CLEAR luciferase reporter activity at 48 hours is increased in Tsc1-cKO (H) and Tsc1-cre (I) keratinocytes, compared with controls. Renilla is used to normalize for luciferase activity. (r = 3; error bars represent SD; P values by Student’s t test). (J) Tsc1-cre keratinocytes transfected with TFEB, TFE3, and MiTF siRNA show increased EGFR and HER2 expression, compared with negative control siRNA, by immunoblot analyses. MITF was immunoblotted separately using the same biological replicate.
Inhibition of hyperactive AKT in mTORC1-inhibited cells rescues autophagy/lysosomal biogenesis and downregulates EGFR expression. Taken together, these data indicate that prolonged increases in mTORC1 activity upregulate lysosomal gene/protein expression via increased total and nuclear levels of MiTs and concomitant CLEAR promoter activity. Similarly, genetic or longer-term pharmacologic mTORC1 inhibition decreased MiT levels and activity and lysosomal gene transcription. Yet these data are at odds with current models suggesting that mTORC1 suppression promotes MiT/TFE transcriptional activity (2–4). One mechanism for this apparent disconnect could be attributed to the activation of alternate signaling pathways bypassing mTORC1 or feedback loops downstream of mTORC1. For example Tsc2-deficient primary neurons showed increased autolysosome formation and autophagic flux via AMPK-dependent ULK1 activation, bypassing mTORC1-dependent ULK1 inhibition (38). Additionally, MiT/TFE activity can be coregulated by numerous kinases, including AKT (15–17). AKT phosphorylation at conserved RXXS/T motifs S467/S565/S510 in TFEB, TFE3, and MITF, respectively, results in their cytoplasmic retention/inactivation (17, 39) or proteasomal degradation (1, 40). We conducted a TMT-based phosphoproteomic analysis of control and Rptor-KO keratinocytes, and found p-TFE3 (S564/565) levels to be significantly increased in Rptor-KO keratinocytes compared with control (Supplemental Figure 8), raising the possibility that AKT was modulating TFE3 activity in the context of Rptor loss. Like mTORC1, AKT can negatively regulate autophagy and lysosomal biogenesis (17, 41). Thus, we tested whether AKT feedback activation in the context of long-term mTORC1 inhibition could inhibit MiT/TFE expression, nuclear localization, and/or activity.
AKT activation was increased in Rheb-cKO and Rptor-cre keratinocytes (Figure 3F, Supplemental Figure 9A, and ref. 31) and decreased in Tsc1-cKO keratinocytes (Figure 2, E and F), consistent with the presence of an mTORC1-AKT feedback loop, as previously documented in other systems (26–28, 42, 43). Accordingly, incubation of Rptor-cre keratinocytes with AKT kinase inhibitors MK2206 (Figure 8A) or GDC-0068 (Supplemental Figure 9B) rescued expression of lysosomal/autophagy target genes, autophagic flux, and MiT/TFE proteins in a dose-dependent manner. MK2206 treatment also the increased the number and perinuclear localization of lysosomes as seen by LAMP1 immunostaining and LysoTracker Red fluorescence (Figure 8, B and C) and increased autophagic vesicles as seen by TEM (Figure 8D). This expansion of the lysosomal/autophagic compartment was driven by MiT/TFE activity since both MK2206 and GDC-0068 stimulated nuclear translocation of MiT/TFE proteins by immunoblot analyses (Figure 8E and Supplemental Figure 9C) and MK2206 also promoted 4X-CLEAR promoter transactivation in luciferase assays (Figure 8F). Correspondingly, MK2206, GDC-0068, and another AKT kinase inhibitor AZD5363 downregulated EGFR and HER2 protein expression in Rptor-cre (Figure 8G and Supplemental Figure 9D) keratinocytes. PI3K inhibition using GDC-0941 in AZD8055-treated keratinocytes (Supplemental Figure 9E) had a similar effect. Genetically silencing AKT1 or AKT2 in Rptor-cre keratinocytes elevated lysosomal proteins and downregulated EGFR, validating the PI3K/AKT inhibitor results (Figure 8H). Conversely, adenoviral-mediated overexpression of constitutively activated myristolated (Myr) AKT1 or AKT2 in Tsc1-cKO keratinocytes downregulated lysosomal proteins and rescued EGFR and HER2 (Figure 8I). These findings confirmed that manipulation of AKT signaling, in the setting of mTORC1 loss or hyperactivity, was sufficient to alter MiT/TFE activity, lysosomal gene expression, and cellular EGFR and HER2 levels.
 Figure 8
Figure 8Inhibition of hyperactive AKT in mTORC1-inhibited cells rescues autophagy/lysosomal biogenesis and downregulates EGFR expression. (A) Immunoblotting showing increased expression of lysosomal, autophagy, and MiT/TFE proteins in Rptor-cre keratinocytes treated with MK2206 (1 μM, 5 μM; 8 hours). Ctsb, LAMP1, and tubulin are noncontemporaneous immunoblots of the same biological replicate, while all other blots are contemporaneous parallel immunoblots of the same biological replicate. (B) LAMP1 immunostaining showing expansion and perinuclear localization of lysosomes in empty and Rptor-cre keratinocytes treated with MK2206 (5 μM, 8 hours), compared with DMSO controls. Scale bar: 50 μm. (C) MK2206-treated Rptor-cre keratinocytes show increased LysoTracker Red fluorescence compared with DMSO controls. Scale bar: 40 μm. (D) Electron micrographs showing increased presence of autophagic vesicles (black arrows) in MK2206-treated Rptor-cre keratinocytes, compared with DMSO controls. Scale bar: 2 μm. (E) MiT/TFE proteins are increased in nuclear-fraction immunoblots of MK2206-treated Rptor-cre keratinocytes (1, 5 μM; 8 hours). Lamin A/C and fibrillarin are used as loading controls. (F) MK2206 treatment of Rptor-cre keratinocytes increases 4X-CLEAR luciferase reporter activity. Renilla is used to normalize for luciferase activity (r = 4; error bars represent SD; P values by Student’s t test). (G) Immunoblotting showing decreased expression of EGFR and HER2 in Rptor-cre keratinocytes treated with MK2206/AZD5363 for 24 hours. (H) Immunoblotting showing increased expression of lysosomal markers and MiT/TFE proteins with downregulation of EGFR expression in Rptor-cre keratinocytes treated with AKT1/2 siRNA. (I) Immunoblotting showing decreased expression of lysosomal markers and MiT/TFE proteins with upregulation of EGFR and HER2 expression in Tsc1-cre keratinocytes infected with Myr-AKT1 or Myr-AKT2 adenovirus.
-
Discussion
To study the effects of constitutive mTORC1 signaling activation or suppression in epithelial tissues, we developed genetically engineered mouse models that allow specific perturbation of mTORC1 signaling in epidermal keratinocytes. We found that mice with epidermal mTORC1 activation developed a phenotype strikingly similar to murine epidermal TGF-α or EGFR loss (20–22). Accordingly, Tsc1 loss in the epidermis suppressed EGFR and HER2 expression and activity, whereas mTORC1 loss-of-function via pharmacological inactivation, or Rheb/Rptor deletion in keratinocytes had the reverse effect. Among all surface receptors, EGFR signaling is prototypically regulated by ligand-induced lysosomal degradation. Significantly, the rate of EGFR decay was significantly enhanced in Tsc1-cKO keratinocytes and decreased in mTORC1-inhibited keratinocytes, confirming that mTORC1 signaling was both necessary and sufficient to regulate the net rate of ligand-induced EGFR degradation.
Lysosomes are critical for the degradation of cellular macromolecules and are transcriptionally regulated by MiT/TFE family members. Interestingly, upregulation of lysosomal function as well as mTORC1 activation are independently essential for epidermal differentiation and barrier function (18, 31). However, the interdependence of mTOR signaling and lysosomal function has not been studied in the skin. Unexpectedly, differential expression analyses comparing epidermis from WT and Rptor-cKO mice revealed that lysosomal genes were significantly downregulated with mTORC1 loss-of-function in the epidermis. Accordingly, multiple lysosomal and autophagy CLEAR genes were upregulated with Tsc1 loss and downregulated with mTORC1 loss-of-function. Probing further upstream, mTORC1 signaling was required to increase the expression, nuclear localization, and transcriptional activity of MiTs, with a global increase in lysosomal content resulting in EGFR and HER2 downregulation.
Our finding that mTORC1 signaling was required to activate lysosomal biogenesis was initially unexpected given the previously established role of mTORC1 as a short-term negative regulator of MiT/TFE-driven lysosomal biogenesis (2–6). We cannot exclude that our results may differ from previously published work in part because our studies used nonimmortalized primary cells exclusively, whereas other studies were predominantly performed in murine embryonic fibroblasts. However, a number of different lines of evidence have since emerged to suggest that the role of mTORC1 in lysosomal gene regulation is likely much more complex than previously thought. First, though initial studies performed in a limited number of cell lines showed that MiT/TFE activity was increased following short-term mTORC1/2 inactivation with Torin1, short-term rapamycin did not affect TFEB phosphorylation or subcellular localization (3). While this could represent rapamycin-insensitive functions of mTORC1, the long-term effects of pharmacological mTORC1 inactivation have not been described thus far. We now show that Torin1 treatment in excess of 24 hours significantly decreases MiT/TFE transcriptional activity. In support of our findings, one previous study reported that temporal mTORC1 inactivation by Torin1 activated TFEB for a limited duration of 1.5 hours, following which it was progressively inactivated (44). Second, previous studies did not directly assess the impact of genetic or constitutive mTORC1 inactivation on lysosomal gene expression. We now show in an unbiased screen via GSEA, that mTORC1 loss-of-function in Rptor-cKO epidermis downregulates multiple lysosomal genes bearing CLEAR regulatory motifs. Finally, in the context of Tsc1/2 loss, several studies have suggested that constitutive activation of mTORC1 paradoxically positively regulates MiT/TFE localization and activity, though the mechanism was not elucidated. Pena-Llopis et al. (11) showed that mTORC1 drives TFEB-dependent V-ATPase gene expression, further reinforcing their findings by publicly available gene expression data sets. Similarly, Tsc1/2 loss promoted TFE3 nuclear localization in ESC cells, Eker rat kidneys, and teratomas in an mTORC1-dependent manner (12, 13).
How can we begin to reconcile these apparently conflicting data? Significantly, MiT/TFE localization is regulated by multiple mTORC1-independent signaling pathways, including ERK and AKT. ERK2-mediated phosphorylation of TFEB at S142 and AKT-mediated phosphorylation at S467/S565/S510 both promote cytosolic MiT/TFE retention and/or degradation (15, 17, 39, 40). Though these previous studies focused on the role of AKT in phosphorylation and nuclear translocation of MiT/TFE proteins, it is notable that we found both total and nuclear levels of MiT/TFEs to be proportionally affected by mTORC1 or AKT modulation. Thus, feedback activation of PI3K/AKT/MAPK signaling following long-term mTORC1 inhibition (25–27) could potentially restrain MiTs, competing with the direct effects of mTORC1. This could also explain why short-term Torin1 treatment promotes mTORC1-inhibited MiT/TFE nuclear localization (by also inhibiting AKT S473/T308 phosphorylation), whereas long-term treatment restrains mTORC1-inhibited MiT/TFE nuclear localization (via feedback upregulation of AKT T308 phosphorylation) (43).
Consistent with this model, we observed the presence of an mTORC1-AKT negative feedback loop in epidermal keratinocytes. What factors could be driving feedback activation of AKT in the context of epidermal Rheb or Rptor loss? There are a number of known signaling intermediates (IRS-1, GRB10) and RTKs (HER3, IGFR, c-MET, PDGFR) which mediate this negative feedback signaling downstream of mTORC1 activity (25–29, 42, 43). Here, we show that EGFR signaling is itself activated downstream of AKT signaling via a decrease in MiT/TFE-mediated lysosomal biogenesis, further reinforcing this negative feedback to AKT. Accordingly, genetic and pharmacological inhibition of AKT in the context of Rheb or Rptor loss completely rescued MiT/TFE-driven transcriptional activity, lysosomal biogenesis, and downregulated EGFR and HER2, while overexpression of constitutively activated AKT in Tsc1-cKO keratinocytes downregulated lysosomal proteins and rescued EGFR and HER2. Notably, EGFR activation can independently trigger lysosomal dysfunction and mimic lysosomal storage diseases (45), potentially synergizing with hyperactive AKT in the context of mTORC1 loss-of-function.
Another unexpected finding in our study is that mTORC1 signaling perturbation modulated lysosomal biogenesis by effects on MiT/TFE levels. While most previous studies have suggested that MiT/TFE activity is regulated via phosphorylation-mediated changes in nuclear-cytoplasmic distribution, several lines of evidence support that altering total levels of these proteins is sufficient to modulate their activity. Gene rearrangements or gene amplifications involving TFEB or MITF result in massive overexpression and constitutive nuclear localization of the full-length TFEB/MITF protein in tumor cells, and a similar mechanism occurs in TFE3-rearranged tumors (46). Though we focused on the study of primary, nontransformed epithelial cells, we observed that in addition to nuclear levels, total MiT/TFE protein levels were dramatically upregulated with Tsc1 loss in an mTORC1-sensitive manner and downregulated with mTORC1 loss-of-function. AKT inhibition fully restored MiT/TFE transcriptional activity and concurrently elevated total levels of MiT/TFE proteins.
What are the potential mechanisms underlying mTORC1-mediated MiT/TFE protein expression and/or turnover? It is known that cellular protein degradation is performed by 2 major systems, the autophagy-lysosome system and the ubiquitin-proteasome system (UPS). These are interactive and compensatory, wherein impairment of one upregulates the activity of the other (47). The UPS carries out degradation of both short-lived regulatory and misfolded proteins, and long-lived ones that form the bulk of the cell (48). However, the role of mTORC1 in the regulation of UPS proteolysis is complex and contextual, since both mTORC1 inhibition (49, 50) and activation (51) can upregulate proteasomal activity. One consequence of lysosomal/autophagy flux defects, seen in many lysosomal storage diseases, is the accumulation of poly-ubiquitinated proteins (52). Interestingly, AKT activation can increase the ubiquitination and proteasomal degradation of specific substrates (53, 54) via phosphorylation, and MiT/TFE proteins are known bonafide proteasomal substrates (55, 56) that can be targeted for degradation by phosphorylation via multiple kinases including AKT (40). Additionally, certain ubiquitin ligases involved in MiT/TFE regulation, such as STUB1 (56), are also responsible for the degradation of substrates in a PI3K/AKT-dependent manner (54). Further studies are required to determine the role of the UPS in MiT/TFE gene regulation, specific ubiquitin ligases or deubiquitinating enzymes (DUBs) modulating MiT/TFE turnover, the phospho-specific residues involved in this process, and the role of PI3K/AKT signaling in mediating this effect.
In addition to enhancing our understanding of the role of mTORC1 signaling in the regulation of lysosomal biogenesis, our data have implications for keratinocyte differentiation and inflammatory skin disorders where mTOR signaling is frequently activated. There are several studies indicating that autophagy and lysosomal function are required for epidermal maturation (18, 57). Thus, the downregulation of MiT/TFE expression and activity with mTORC1 loss-of-function may contribute to the keratinocyte differentiation and skin barrier defect we observed in previous work in the Rptor-cKO mice (31). Accordingly, lysosomal dysfunction may also interfere with lipid biosynthesis, contributing to human skin disorders with defective barrier function such as atopic dermatitis (58). In other inflammatory disorders such as psoriasis, mTORC1 signaling is hyperactivated (59) and lysosomal function or dysfunction may contribute to aberrant epidermal homeostasis (60). In future studies, it will be of interest to investigate how changes in lysosomal biology downstream of mTORC1 signaling may be mechanistically important and potential therapeutic targets in inflammatory skin disease.
-
Methods
Mice
Animal protocols were approved by the Johns Hopkins University Animal Care and Use Committee. The following strains were used: mice expressing cre recombinase under control of the human K14 promoter (KRT14-cre) (stock number 004782, STOCK Tg(KRT14-cre)1Amc/J), mice carrying loxP sites flanking exons 17 and 18 of Tsc1 (stock number 005680, Tsc1tm1Djk/J), mice carrying loxP sites flanking exons 2, 3, and 4 of Tsc2 (stock number 027458, Tsc2tm1.1Mjgk/J), and mice carrying loxP sites flanking exon 6 of Rptor (stock number 013188, B6Cg-Rptortm1.1Dmsa/J) were from The Jackson Laboratory; mice with loxP-flanked Rheb S16H alleles and mice with loxP-flanked Rheb1 alleles were generated in the laboratory of P.F. Worley (Department of Neuroscience, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA).
Epidermal-specific deletion of Tsc1 or transgenic expression of Rheb S16H was obtained by crossing homozygously expressing KRT14-cre mice with Tsc1 or S16Hfl/fl mice. Epidermal-specific deletion of Rheb or Rptor was obtained by crossing hemizygously expressing KRT14-cre mice with Rhebfl/fl or Rptorfl/fl mice. All experiments were performed on E18.5–E19.5 embryos and P0–P7 pups. Genomic DNA was isolated from tail snips and genotyping performed using the following primers: WT and floxed Tsc1: 5′-GAATCAACCCCACAGAGC AT-3′ (forward), 5′-GTCACGACCGTAGGAGAAGC-3′ (reverse); floxed S16H: 5′-GCAACGTGCTGGTTATTGTG-3′ (forward), 5′-GGGGAACTTCCTGACTAGGG-3′ (reverse); excised S16H: 5′-CAGCCATTGCCTTTTATGGT-3′ (forward), 5′-ACCACCACCACCATTGAGAT-3′ (reverse); WT and floxed Rptor: 5′-CTCAGTAGTGGTATGTGCTCAG-3′ (forward), 5′-GGGTACAGTATGTCAGCACAG-3′ (reverse); WT and floxed Rheb1: 5′-GCCCAGAACATCTGTTCCAT-3′ (forward), 5′-GGTACCCACAACCTGACACC-3′ (reverse); recombined Rheb1: 5′-ATAGCTGGAGCCACCAACAC-3′ (forward), 5′-GCCTCAGCTTCTCAAGCAAC-3′ (reverse); KRT14-cre: 5′-TTCCTCAGGAGTGTCTTCGC-3′ (transgene), 5′-GTCCATGTCCTTCCTGAGC-3′ (transgene), 5′-CAAATGTTGCTTGTCTGGTG-3′ (internal positive control forward), 5′-GTCAGTCGAGTGCACAGTTT-3′ (internal positive control reverse).
Primary mouse keratinocyte cultures
Primary mouse keratinocytes were isolated from newborn (P0/P7) skin. Newborn pups were decapitated, immersed in 7.5% povidone-iodine for 5 minutes, and rinsed in 70% ethanol for 2 minutes. The trunk skin was removed and placed dermis-side down in a Petri dish containing 0.25% trypsin-EDTA (Invitrogen) overnight at 4°C for 18 hours. The dermis was separated from the epidermis and keratinocytes isolated by scraping the basal surface of the epidermis. Keratinocyte cell suspensions were passed through a 100-μm cell strainer, centrifugated twice, and plated on Petri dishes coated with fibronectin (F1141; MilliporeSigma), in mouse keratinocyte medium (mKer) containing the following ingredients for a final volume of 500 mL: 3 parts low glucose DMEM (337.5 mL); 1 part Ham’s F-12 (112.5 mL); 10% FBS (50 mL); penicillin (60 μg/mL) (1 mL of 104 U/mL stock); gentamycin (25 μg/mL) (250 μL of 50 mg/mL stock); insulin (5 μg/mL) (250 μL of 10 mg/mL stock); hydrocortisone (0.4 μg/mL) (200 μL of 1 mg/mL stock); cholera toxin (10–10 M) (5 μL of 1 mg/mL stock); transferrin (5 μg/mL) + 3,3-5′triiodo-L-thyronine (T3) (2 × 10–9 M) (500 μL of a T3-transferrin stock).
To obtain keratinocytes with genetic ablation of Rptor or Tsc1, Rptorfl/fl or Tsc1fl/fl keratinocytes were infected with Cre-recombinase–expressing or empty adenoviral vectors (Vector Biolabs), prior to plating cells.
Reagents and antibodies
Primary antibodies. The following antibodies were used: Tsc1 (catalog 6935, Cell Signaling), 1:1000; Tsc2 (catalog 3990, Cell Signaling), 1:1000; Rheb (catalog 09-247, Millipore), 1:1000; Raptor (catalog 2280, Cell Signaling), 1:1000; Raptor (catalog 05-1470, MilliporeSigma), 1:400; Phospho-S6 Ribosomal Protein (Ser240/244) (catalog 5364, Cell Signaling), 1:800-1:1000; S6 Ribosomal Protein (catalog 2317, Cell Signaling), 1:1000; Phospho-4E BP1 (T37/46) (catalog 2855, Cell Signaling), 1:1000; 4E-BP1 (catalog 9644, Cell Signaling), 1:1000; Phospho-p70 S6 Kinase (T37/46) (catalog 9205, Cell Signaling), 1:1000; p70 S6 Kinase (catalog 9202, Cell Signaling), 1:1000; β-Actin (catalog 3700, Cell Signaling), 1:4000; Gapdh (catalog 2118, Cell Signaling), 1:4000; EGFR (catalog sc-03, Santa Cruz), 1:500; HER2 (catalog sc-284, Santa Cruz), 1:500; p-EGFR (Y1068) (catalog 2234, Cell Signaling), 1:250; Phospho-Erk1/2 (catalog 9101, Cell Signaling), 1:1000; Erk1/2 (catalog 4695, Cell Signaling), 1:1000; Phospho-Akt (S473) (catalog 4060, Cell Signaling), 1:1000; Phospho-Akt (T308) (catalog 5106, Cell Signaling), 1:500; Akt (pan) (catalog 4691, Cell Signaling), 1:1000; Phospho-FoxO1 (Thr24)/FoxO3a (Thr32)/FoxO4 (Thr28) (catalog 2599, Cell Signaling), 1:1000; FoxO1 (catalog 2880, Cell Signaling), 1:1000; Na,K-ATPase (catalog 3010, Cell Signaling), 1:1000; LAMP-1 (catalog sc-19992, Santa Cruz), 1:500; LAMP-2 (catalog ABL-93, Developmental Studies Hybridoma Bank at the University of Iowa), 1:50; CTSB (catalog 31718, Cell Signaling), 1:1000; CTSD (catalog sc-6486, Santa Cruz), 1:500; Rab7 (catalog 9367, Cell Signaling), 1:1000; LAMTOR1 (catalog 8975, Cell Signaling), 1:1000; LAMTOR2 (catalog 8145, Cell Signaling), 1:1000; LAMTOR3 (catalog 8168, Cell Signaling), 1:1000; RagA (catalog 4357, Cell Signaling), 1:1000; RagB (catalog 8150, Cell Signaling), 1:1000; RagC (catalog 5466, Cell Signaling), 1:1000; LAPTM4B (catalog ABC290, EMD Millipore), 1:1000; p62/SQSTM1 (catalog 23214, Cell Signaling), 1:1000; ATP6AP2 (catalog 10926-1-1AP, Proteintech), 1:500; ATP6V0A1 (catalog sc-374475, Santa Cruz), 1:500; ATP6v1b1b2 (catalog sc-55544, Santa Cruz), 1:500; ATP6v1c1 (catalog sc-271077, Santa Cruz), 1:500; ATP6v1d (catalog sc-166218, Santa Cruz), 1:500; Atg3 (catalog 3415, Cell Signaling), 1:1000; Atg5 (catalog 12994, Cell Signaling), 1:1000; Atg7 (catalog 8558, Cell Signaling), 1:1000; Atg16L1 (catalog 8089, Cell Signaling), 1:1000; TFEB (catalog A303-673A, Bethyl), 1:500; TFE3 (catalog PA5-54909, Thermo Fisher Scientific), 1:500; TFE3 (catalog ABE1400, MilliporeSigma), 1:500; MITF (catalog 13092-1-1AP, Proteintech), 1:500; MITF (catalog 12590, Cell Signaling), 1:500; Histone H3 (catalog 4499, Cell Signaling), 1:1000; Fibrillarin (catalog 2639, Cell Signaling), 1:1000; Lamin A/C (catalog 4777, Cell Signaling), 1:1000; Beclin-1 (catalog 3495, Cell Signaling), 1:1000; LC3A (catalog 4599, Cell Signaling), 1:1000.
Reagents. DMEM (catalog 11885084, Thermo Fisher Scientific), Ham’s F-12 (catalog 11765054, Thermo Fisher Scientific), EGF (Peprotech), FBS (Hyclone), T3/transferrin (MilliporeSigma), hydrocortisone and cholera toxin (MilliporeSigma), insulin (Roche), gentamycin (Amresco), Mg2+ lysis/wash buffer (catalog 20-168, MilliporeSigma), 8M Urea (Amresco), cell lysis buffer (catalog 9803, Cell Signaling), rapamycin and AZD8055 (LC Laboratories), MK2206, GDC-0068, AZD8186, and GDC-0941 (Selleckchem), Torin1 and AKT1, 2 SignalSilence siRNA (Cell Signaling), Silencer Select Negative Control siRNA, Lipofectamine 3000 reagent, Lipofectamine RNAiMAX reagent LysoTracker DND-99 (Thermo Fisher Scientific), siGENOME Mouse siRNA SMARTpool (TFEB, TFE3, and MITF; Dharmacon), Adeno CMV Null, Cre Recombinase, Akt1 (Myr) and Akt2 (Myr) adenoviruses (Vector Biolabs), Magic Red Cathepsin B Kit (catalog ICT937, Biorad). 4XCLEAR-luciferase reporter was a gift from Albert La Spada (Addgene plasmid 66880) (37).
Histology and immunostaining
Mouse skins were fixed in 10% neutral buffered formalin (Sigma-Aldrich), embedded in paraffin, sectioned at 4 μm, and used for immunohistochemistry. Sections were deparaffinized in xylene (Sigma-Aldrich), hydrated in graded ethanol, and rinsed in distilled water. Antigen retrieval was performed using citrate (10 mM, pH 6.0) or EDTA + 0.01% Tween 20 (1 mM, pH 8.0) buffers and the HIER (heat-induced epitope retrieval) method, in accordance with the protocol specified for each antibody. All washing steps were done using 1X TBS-T buffer. Endogenous peroxidase activity was quenched by incubation with Dual Enzyme Block (Dako, Agilent Technologies) for 10 minutes at room temperature. Sections were incubated with each antibody overnight at 4°C diluted in antibody dilution buffer (Roche/Ventana Medical Systems). For immunohistochemistry, a horseradish peroxidase–labeled polymer, Poly-HRP PowerVision Detection System (Novocastra/Leica Biosystems), was applied for 30 minutes at room temperature. Signal detection was performed using DAB (Sigma-Aldrich) for 20 minutes at room temperature. Slides were counterstained for 30 seconds with Mayer’s hematoxylin (Dako, Agilent Technologies), dehydrated, and mounted. For immunofluorescence, after primary antibody overnight reaction at 4°C, sections were incubated with secondary antibodies (Alexa Fluor 488– or Alexa Fluor 594–conjugated, anti–rabbit or anti–mouse IgG; Thermo Fisher Scientific) at a dilution of 1:200 for 1 hour 30 minutes at room temperature. Subsequently, they were washed 2 times for 5 minutes each time in PBS, rinsed in distilled water, dehydrated in graded ethanol, and mounted with ProLong Gold Antifade with DAPI (Thermo Fisher Scientific). IHC for TFE3 was carried out as previously described (61).
Protein lysate preparation and immunoblotting
Mouse epidermis was separated from the dermis following incubation of pup skin with 3.8% ammonium thiocyanate (catalog A7149, Sigma-Aldrich) for 10 minutes at room temperature. The epidermal sheet was homogenized using gentleMACS M tubes in the gentleMACS dissociator (Miltenyi Biotec). Tissues or cells were homogenized and lysed in ice-cold 1X Mg2+ lysis/wash buffer (catalog 20-168, MilliporeSigma) or RIPA buffer (catalog R0278, MilliporeSigma) supplemented with NaVO4 (1 mM), NaF (1 mM), and 10 μL Halt Protease and Phosphatase Inhibitor Cocktail (catalog 78440, Thermo Fisher Scientific) in 1 mL buffer for 15 minutes on ice. Lysates were sheared by passing through 20-, 22-, 25-, and 26-gauge needles progressively, centrifuged at 21,300 g for 10 minutes at 4°C, and supernatants collected. Protein concentrations were quantified using the BCA Protein Assay Kit (catalog 23225, Pierce), and 5–10 μg protein was resolved on a 1.5-mm, 3%–8% Tris-Acetate or 4%–12% Bis-Tris SDS-PAGE gel (Thermo Fisher Scientific). Protein was transferred to nitrocellulose membranes (Amersham Bioscience). Membranes were allowed to block for 1 hour at room temperature in 5% nonfat milk in 1X TBS-T and then incubated overnight with a primary antibody diluted in 5% BSA in 1X TBS-T. The secondary antibodies used were anti–rabbit or anti–mouse Ig as appropriate (Cell Signaling) and diluted at 1:1000 in 5% nonfat milk in 1X TBS-T. Blots were developed using a chemiluminescent development solution (Super Signal West Femto, Pierce) and bands were imaged on a chemiluminescent imaging system (ChemiDoc Touch imaging System, Biorad) or MicroChemi Chemiluminescent Imager (FroggaBio Inc.). Digital images were quantified using background correction on the Alpha Innotech system (Protein Simple) and all bands were normalized to their respective β-actin, tubulin, or GAPDH expression levels as loading controls. Nuclear lysates were prepared using the PARIS kit (catalog AM1921, Thermo Fisher Scientific) according to manufacturer’s instructions. Digital images were quantified using background correction on the Alpha Innotech system and all bands were normalized to their respective lamin, histone H3, or fibrillarin levels as loading controls. Cell surface biotinylation was performed using the Pierce Cell Surface Protein Isolation Kit (catalog 89881, Thermo Fisher Scientific) according to the manufacturer’s instructions. Digital images were quantified using background correction on the Alpha Innotech system and all bands were normalized to their respective Na/K-ATPase levels as loading controls. Statistical analysis was performed using Student’s unpaired t test.
siRNA-mediated gene silencing and plasmid transfection
Primary mouse keratinocytes were transfected with 50 nm siRNA using Lipofectamine RNAiMAX reagent using the reverse transfection protocol according to the transfection guidelines. Primary keratinocytes were transfected with cDNA constructs using Lipofectamine 3000 reagent (L3000008, Thermo Fisher Scientific) according to the transfection guidelines.
RNA isolation and quantitative real-time RT-PCR
Total cellular RNA was extracted using either TRIzol (catalog 15596026, Invitrogen) for epidermal tissue, or RNeasy Mini kit (catalog 74104, Qiagen) for keratinocytes according to manufacturer’s instructions. RNA was converted to cDNA using SuperScript III First-Strand Synthesis System (catalog 18080051, Thermo Fisher Scientific) according to manufacturer’s instructions. mRNA levels were quantified using an ABI Prism 7900HT Real-Time PCR system (Applied Biosystems) with the following primers and probes: ATP6AP2(Mm00510396_m1), ATP6V0A (Mm00441838_m1), ATP6V0B (Mm00504328_m1), ATP6V1A (Mm01343719_m1), ATP6V1B2 (Mm00431987_m1), ATP6V1C2 (Mm00505047_m1), ATP6V1D (Mm00445832_m1), ATP6V1E1 (Mm00657610_m1), LAMP1 (Mm00495262_m1), CTSB (Mm01310506_m1), CTSD (Mm00515586_m1), CTSK (Mm00484039_m1), MCOLN1 (Mm00522550_m1), SQSTM1 (Mm00448091_m1), TFEB (Mm00448968_m1), TFE3 (Mm01341186_m1), MITF (Mm00434954_m1), EGFR (Mm01187858_m1), ERBB2 (Mm00658541_m1), ACTB (Mm02619850_m1). Threshold cycle (Ct) was obtained from the PCR reaction curves and mRNA levels were quantitated using the comparative Ct method with actin mRNA serving as the reference. Statistical analysis was performed using Student’s unpaired t test.
Lysosomal expression and activity assays
Lysosomal fractionation assays. Lysosomal fractionation assays were carried out as previously described (62). Cultured keratinocytes grown on 150 mm dishes were harvested and lysed in 750 mL of cold fractionation buffer (50 mM KCl, 90 mM potassium gluconate, 1 mM EGTA, 50 mM sucrose, 5 mM glucose, protease inhibitor cocktail tablet, and 20 mM HEPES, pH 7.4). The cells were then lysed by syringing, and nuclear fraction was removed by centrifugation at 1000g for 10 minutes at 4°C. The supernatant was then centrifuged at 20,000g for 30 minutes at 4°C. The precipitated lysosome-enriched fraction (LEF) was resuspended in the fractionation buffer, and the supernatant was separated as the cytosolic fraction.
Cathepsin B activity assays. To measure lysosomal cathepsin B activity, cells were incubated with Magic Red Cathepsin B (Biorad) for 1 hour and processed according to the manufacturer’s instructions for fluorescence plate reader analysis.
Immunocytochemistry
Primary mouse keratinocytes were seeded on coverslips coated with fibronectin. Following experimental treatments, cells were either fixed in 100% methanol at –20°C for 30 minutes or 4% PFA for 15 minutes at room temperature, according to antibody specifications. Following 3 rinses in 1X PBS, cells were permeabilized and blocked in a buffer containing 1X PBS, 5% normal donkey serum, and 0.3% Triton X-100. For immunofluorescence, coverslips were incubated with the indicated primary antibodies overnight at 4°C in antibody dilution buffer (ADB) containing 1X PBS, 1% BSA, and 0.3% Triton X-100. After 3 rinses of 1X PBS, coverslips were incubated with secondary antibodies (Alexa Fluor 488– or Alexa Fluor 594–conjugated, anti–rabbit or anti–mouse IgG; Thermo Fisher Scientific) in ADB at a dilution of 1:200 for 1 hour at room temperature. Nuclei were counterstained with DAPI and coverslips visualized using an Olympus BX41 epifluorescence microscope.
Immunofluorescence image analysis and quantification
Image analysis and quantification were done in ImageJ.
Lamp1 immunostaining and quantification. Confocal images were acquired on a Nikon TE-2000e microscope and using the NIS elements 5.0.1 software. All images were captured using the same exposure and gain settings followed by automatic deconvolution. The area of Lamp1 was measured using Image J and normalized to the number of nuclei.
Quantification of nuclear TFE3 intensity. Cells were stained with DAPI to mark nuclei (blue channel) and anti-total TFE3 (red channel). Images were analyzed using ImageJ. The blue channel was used to segment nuclei as follows: images were thresholded to remove background and converted to binary images, following which the Analyze Particles function was used for automatic detection of nuclear outlines. These nuclear outlines were applied to the red channel and mean fluorescence intensity of TFE3 within the regions was measured.
Statistics for image analysis. Normal distribution was assessed using the D’Agostino & Pearson normality test. If normally distributed, statistical significance was determined with Student’s t test when comparing 2 experimental groups, or with 1-way ANOVA with Dunnett’s correction when comparing 3 or more experimental groups. If not normally distributed, statistical significance was determined with the Mann-Whitney test when comparing 2 experimental groups, or with the Kruskal-Wallis test with Dunn’s correction when comparing 3 or more experimental groups. All tests assumed a 2-tailed deviation and were performed in Prism 7 (GraphPad).
Transmission electron microscopy
Mouse skin and keratinocyte samples were fixed in 2.5% glutaraldehyde, 3 mM MgCl2 and 1% sucrose, in 0.1M sodium cacodylate buffer, pH 7.2 at 4°C overnight, followed by 3 buffer rinses, 15 minutes each, in 3 mM MgCl2, 3% sucrose, and 0.1M sodium cacodylate. The samples were postfixed in 1% osmium tetroxide in 0.1M sodium cacodylate for 1 hour on ice in the dark, rinsed twice with distilled water for 5 minutes, stained with 2% aqueous uranyl acetate (0.22 μm filtered) for 1 hour in the dark, followed by dehydration in an ascending grade of ethanol (50%, 70%, 90%, and 100%; thrice each), and embedded in an epoxy resin. The resin was allowed to polymerize at 37°C overnight for 2–3 days followed by 60°C overnight. Grids were stained with 2% uranyl acetate in 50% methanol, followed by lead citrate, and observed with a Philips CM120 at 80 kV. Images were captured with an AMT XR80 high-resolution (16-bit) 8 megapixel camera.
Microarray analysis
Microarray-based differential expression analysis of E18.5 epidermis from WT/Rptor-cKO mice was carried out as described in Supplemental Methods. All original microarray data were deposited in the NCBI’s Gene Expression Omnibus database (GEO 124754).
Statistics
For image analysis, RNA and protein quantification and luciferase assays, statistical significance was determined using the unpaired, 2-tailed Student’s t test when comparing 2 experimental groups, or with 1-way ANOVA with Tukey’s correction when comparing 3 or more experimental groups. All tests were performed in Prism 8 (GraphPad). P values of less than 0.05 were considered statistically significant.
Phosphoproteome analysis
TMT-based phosphoproteome analysis of control or Rptor-KO mouse primary keratinocytes was carried out as described in Supplemental Methods.
-
Author contributions
AS, KA, and TLL conceived and designed the study. KA, AS, SM, CHN, HK, ZK, MN, MS, BL, CCT, BS, and TLL acquired data. KA, SM, CHN, AS, RKA, CCT, BS, MS, and TLL analyzed data. KA, SM, PP, CCT, and TLL drafted the manuscript.
-
Acknowledgments
The authors thank Avi Rosenberg for assistance with analyzing TEM images and Paul Worley for providing us the Rhebfl/fl and Rheb S16Hfl/fl mice. This work was funded by the NIH (R01 CA200858-01).
Address correspondence to: Tamara Lotan and Kaushal Asrani, 1550 Orleans Street, Baltimore, Maryland 21231, USA. Phone: 410.614.9196; Email: tlotan1@jhmi.edu, kasrani1@jhmi.edu.
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(12):5584–5599.https://doi.org/10.1172/JCI128287.
-
References
- Puertollano R, Ferguson SM, Brugarolas J, Ballabio A. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 2018;37(11):e98804. View this article via: PubMed Google Scholar
- Roczniak-Ferguson A, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5(228):ra42. View this article via: PubMed Google Scholar
- Settembre C, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31(5):1095–1108.
- Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8(6):903–914.
- Martina JA, et al. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal. 2014;7(309):ra9.
- Napolitano G, et al. mTOR-dependent phosphorylation controls TFEB nuclear export. Nat Commun. 2018;9(1):3312.
- Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–976.
- Yu L, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465(7300):942–946.
- Baar K, Esser K. Phosphorylation of p70(S6k) correlates with increased skeletal muscle mass following resistance exercise. Am J Physiol. 1999;276(1):C120–C127.
- Mansueto G, et al. Transcription factor EB controls metabolic flexibility during exercise. Cell Metab. 2017;25(1):182–196.
- Peña-Llopis S, et al. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 2011;30(16):3242–3258.
- Betschinger J, Nichols J, Dietmann S, Corrin PD, Paddison PJ, Smith A. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell. 2013;153(2):335–347.
- Kawano H, et al. Aberrant differentiation of Tsc2-deficient teratomas associated with activation of the mTORC1-TFE3 pathway. Oncol Rep. 2015;34(5):2251–2258.
- Di Malta C, et al. Transcriptional activation of RagD GTPase controls mTORC1 and promotes cancer growth. Science. 2017;356(6343):1188–1192.
- Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–1433.
- Li Y, et al. Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat Cell Biol. 2016;18(10):1065–1077.
- Palmieri M, et al. mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat Commun. 2017;8:14338. View this article via: PubMed Google Scholar
- Monteleon CL, et al. Lysosomes Support the Degradation, Signaling, and Mitochondrial Metabolism Necessary for Human Epidermal Differentiation. J Invest Dermatol. 2018;138(9):1945–1954.
- Kobayashi T, et al. A germ-line Tsc1 mutation causes tumor development and embryonic lethality that are similar, but not identical to, those caused by Tsc2 mutation in mice. Proc Natl Acad Sci USA. 2001;98(15):8762–8767.
- Luetteke NC, Qiu TH, Peiffer RL, Oliver P, Smithies O, Lee DC. TGF alpha deficiency results in hair follicle and eye abnormalities in targeted and waved-1 mice. Cell. 1993;73(2):263–278.
- Lichtenberger BM, et al. Epidermal EGFR controls cutaneous host defense and prevents inflammation. Sci Transl Med. 2013;5(199):199ra111. View this article via: PubMed Google Scholar
- Schneider MR, Werner S, Paus R, Wolf E. Beyond wavy hairs: the epidermal growth factor receptor and its ligands in skin biology and pathology. Am J Pathol. 2008;173(1):14–24.
- Zou J, et al. Rheb1 is required for mTORC1 and myelination in postnatal brain development. Dev Cell. 2011;20(1):97–108.
- Chong-Kopera H, et al. TSC1 stabilizes TSC2 by inhibiting the interaction between TSC2 and the HERC1 ubiquitin ligase. J Biol Chem. 2006;281(13):8313–8316.
- Carracedo A, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118(9):3065–3074.
- Harrington LS, et al. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol. 2004;166(2):213–223.
- Yu Y, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332(6035):1322–1326.
- Zhang H, et al. PDGFRs are critical for PI3K/Akt activation and negatively regulated by mTOR. J Clin Invest. 2007;117(3):730–738.
- Muranen T, et al. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell. 2012;21(2):227–239.
- Wei F, Zhang Y, Geng L, Zhang P, Wang G, Liu Y. mTOR inhibition induces EGFR feedback activation in association with its resistance to human pancreatic cancer. Int J Mol Sci. 2015;16(2):3267–3282.
- Asrani K, et al. mTORC1 loss impairs epidermal adhesion via TGF-β/Rho kinase activation. J Clin Invest. 2017;127(11):4001–4017.
- Goh LK, Sorkin A. Endocytosis of receptor tyrosine kinases. Cold Spring Harb Perspect Biol. 2013;5(5):a017459. View this article via: PubMed Google Scholar
- Longva KE, Blystad FD, Stang E, Larsen AM, Johannessen LE, Madshus IH. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J Cell Biol. 2002;156(5):843–854.
- Sigismund S, et al. Threshold-controlled ubiquitination of the EGFR directs receptor fate. EMBO J. 2013;32(15):2140–2157.
- Perera RM, et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524(7565):361–365.
- Palmieri M, et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet. 2011;20(19):3852–3866.
- Cortes CJ, et al. Polyglutamine-expanded androgen receptor interferes with TFEB to elicit autophagy defects in SBMA. Nat Neurosci. 2014;17(9):1180–1189.
- Di Nardo A, et al. Neuronal Tsc1/2 complex controls autophagy through AMPK-dependent regulation of ULK1. Hum Mol Genet. 2014;23(14):3865–3874.
- Pi H, et al. AKT inhibition-mediated dephosphorylation of TFE3 promotes overactive autophagy independent of MTORC1 in cadmium-exposed bone mesenchymal stem cells. Autophagy. 2019;15(4):565–582.
- Wang C, et al. Phosphorylation of MITF by AKT affects its downstream targets and causes TP53-dependent cell senescence. Int J Biochem Cell Biol. 2016;80:132–142.
- Wang RC, et al. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338(6109):956–959.
- Rodrik-Outmezguine VS, et al. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov. 2011;1(3):248–259.
- Yoon SO, et al. Focal adhesion- and IGF1R-dependent survival and migratory pathways mediate tumor resistance to mTORC1/2 inhibition. Mol Cell. 2017;67(3):512–527.e4.
- Marin Zapata PA, Beese CJ, Jünger A, Dalmasso G, Brady NR, Hamacher-Brady A. Time course decomposition of cell heterogeneity in TFEB signaling states reveals homeostatic mechanisms restricting the magnitude and duration of TFEB responses to mTOR activity modulation. BMC Cancer. 2016;16:355. View this article via: PubMed Google Scholar
- De Pasquale V, et al. EGFR activation triggers cellular hypertrophy and lysosomal disease in NAGLU-depleted cardiomyoblasts, mimicking the hallmarks of mucopolysaccharidosis IIIB. Cell Death Dis. 2018;9(2):40.
- Kauffman EC, et al. Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers. Nat Rev Urol. 2014;11(8):465–475.
- Pandey UB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447(7146):859–863.View this article via: PubMed Google Scholar
- Rock KL, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78(5):761–771.
- Zhao J, Zhai B, Gygi SP, Goldberg AL. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc Natl Acad Sci USA. 2015;112(52):15790–15797.
- Rousseau A, Bertolotti A. An evolutionarily conserved pathway controls proteasome homeostasis. Nature. 2016;536(7615):184–189.
- Zhang Y, et al. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 2014;513(7518):440–443.
- Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–884.
- Aoki M, Jiang H, Vogt PK. Proteasomal degradation of the FoxO1 transcriptional regulator in cells transformed by the P3k and Akt oncoproteins. Proc Natl Acad Sci USA. 2004;101(37):13613–13617.
- Terme JM, Lhermitte L, Asnafi V, Jalinot P. TGF-beta induces degradation of TAL1/SCL by the ubiquitin-proteasome pathway through AKT-mediated phosphorylation. Blood. 2009;113(26):6695–6698.
- Zhao X, Fiske B, Kawakami A, Li J, Fisher DE. Regulation of MITF stability by the USP13 deubiquitinase. Nat Commun. 2011;2:414. View this article via: PubMed Google Scholar
- Sha Y, Rao L, Settembre C, Ballabio A, Eissa NT. STUB1 regulates TFEB-induced autophagy-lysosome pathway. EMBO J. 2017;36(17):2544–2552.
- Yoshihara N, et al. The significant role of autophagy in the granular layer in normal skin differentiation and hair growth. Arch Dermatol Res. 2015;307(2):159–169.
- Elias PM, Wakefield JS. Mechanisms of abnormal lamellar body secretion and the dysfunctional skin barrier in patients with atopic dermatitis. J Allergy Clin Immunol. 2014;134(4):781–791.e1.
- Buerger C, et al. Inflammation dependent mTORC1 signaling interferes with the switch from keratinocyte proliferation to differentiation. PLoS ONE. 2017;12(7):e0180853.
- Akinduro O, et al. Constitutive autophagy and nucleophagy during epidermal differentiation. J Invest Dermatol. 2016;136(7):1460–1470.
- Argani P, et al. A distinctive subset of PEComas harbors TFE3 gene fusions. Am J Surg Pathol. 2010;34(10):1395–1406.
- Kim YC, et al. Rag GTPases are cardioprotective by regulating lysosomal function. Nat Commun. 2014;5:4241. View this article via: PubMed Google Scholar
- Puertollano R, Ferguson SM, Brugarolas J, Ballabio A. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 2018;37(11):e98804.
-
Version history
- Version 1 (September 17, 2019): In-Press Preview
- Version 2 (November 11, 2019): Electronic publication
- Version 3 (December 2, 2019): Print issue publication



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)








