Advertisement
Research ArticleOncologyTherapeutics Free access | 10.1172/JCI122216
Peptide-based PET quantifies target engagement of PD-L1 therapeutics
Dhiraj Kumar,1 Ala Lisok,1 Elyes Dahmane,2 Matthew McCoy,3 Sagar Shelake,1 Samit Chatterjee,1 Viola Allaj,4 Polina Sysa-Shah,1 Bryan Wharram,1 Wojciech G. Lesniak,1 Ellen Tully,5 Edward Gabrielson,5,6 Elizabeth M. Jaffee,5,6 John T. Poirier,4 Charles M. Rudin,4 Jogarao V.S. Gobburu,2 Martin G. Pomper,1,6,7 and Sridhar Nimmagadda1,6,7,8
1The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Center for Translational Medicine, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
3Innovation Center for Biomedical Informatics, Georgetown University, Washington, DC, USA.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
6The Sidney Kimmel Comprehensive Cancer Center and the Bloomberg–Kimmel Institute for Cancer Immunotherapy, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
7Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
8Division of Clinical Pharmacology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Sridhar Nimmagadda, Johns Hopkins Medical Institutions, 1550 Orleans Street, CRB II, Rm 491, Baltimore, Maryland 21287, USA. Phone: 410.502.6244; Email: snimmag1@jhmi.edu.
Authorship note: DK and AL contributed equally to this work.
Find articles by Kumar, D. in: JCI | PubMed | Google Scholar
1The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Center for Translational Medicine, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
3Innovation Center for Biomedical Informatics, Georgetown University, Washington, DC, USA.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
6The Sidney Kimmel Comprehensive Cancer Center and the Bloomberg–Kimmel Institute for Cancer Immunotherapy, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
7Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
8Division of Clinical Pharmacology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Sridhar Nimmagadda, Johns Hopkins Medical Institutions, 1550 Orleans Street, CRB II, Rm 491, Baltimore, Maryland 21287, USA. Phone: 410.502.6244; Email: snimmag1@jhmi.edu.
Authorship note: DK and AL contributed equally to this work.
Find articles by Lisok, A. in: JCI | PubMed | Google Scholar
1The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Center for Translational Medicine, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
3Innovation Center for Biomedical Informatics, Georgetown University, Washington, DC, USA.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
6The Sidney Kimmel Comprehensive Cancer Center and the Bloomberg–Kimmel Institute for Cancer Immunotherapy, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
7Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
8Division of Clinical Pharmacology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Sridhar Nimmagadda, Johns Hopkins Medical Institutions, 1550 Orleans Street, CRB II, Rm 491, Baltimore, Maryland 21287, USA. Phone: 410.502.6244; Email: snimmag1@jhmi.edu.
Authorship note: DK and AL contributed equally to this work.
Find articles by Dahmane, E. in: JCI | PubMed | Google Scholar
1The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Center for Translational Medicine, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
3Innovation Center for Biomedical Informatics, Georgetown University, Washington, DC, USA.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
6The Sidney Kimmel Comprehensive Cancer Center and the Bloomberg–Kimmel Institute for Cancer Immunotherapy, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
7Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
8Division of Clinical Pharmacology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Sridhar Nimmagadda, Johns Hopkins Medical Institutions, 1550 Orleans Street, CRB II, Rm 491, Baltimore, Maryland 21287, USA. Phone: 410.502.6244; Email: snimmag1@jhmi.edu.
Authorship note: DK and AL contributed equally to this work.
Find articles by McCoy, M. in: JCI | PubMed | Google Scholar
1The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Center for Translational Medicine, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
3Innovation Center for Biomedical Informatics, Georgetown University, Washington, DC, USA.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
6The Sidney Kimmel Comprehensive Cancer Center and the Bloomberg–Kimmel Institute for Cancer Immunotherapy, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
7Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
8Division of Clinical Pharmacology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Sridhar Nimmagadda, Johns Hopkins Medical Institutions, 1550 Orleans Street, CRB II, Rm 491, Baltimore, Maryland 21287, USA. Phone: 410.502.6244; Email: snimmag1@jhmi.edu.
Authorship note: DK and AL contributed equally to this work.
Find articles by
Shelake, S.
in:
JCI
|
PubMed
|
Google Scholar
|

1The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Center for Translational Medicine, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
3Innovation Center for Biomedical Informatics, Georgetown University, Washington, DC, USA.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
6The Sidney Kimmel Comprehensive Cancer Center and the Bloomberg–Kimmel Institute for Cancer Immunotherapy, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
7Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
8Division of Clinical Pharmacology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Sridhar Nimmagadda, Johns Hopkins Medical Institutions, 1550 Orleans Street, CRB II, Rm 491, Baltimore, Maryland 21287, USA. Phone: 410.502.6244; Email: snimmag1@jhmi.edu.
Authorship note: DK and AL contributed equally to this work.
Find articles by Chatterjee, S. in: JCI | PubMed | Google Scholar
1The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Center for Translational Medicine, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
3Innovation Center for Biomedical Informatics, Georgetown University, Washington, DC, USA.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
6The Sidney Kimmel Comprehensive Cancer Center and the Bloomberg–Kimmel Institute for Cancer Immunotherapy, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
7Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
8Division of Clinical Pharmacology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Sridhar Nimmagadda, Johns Hopkins Medical Institutions, 1550 Orleans Street, CRB II, Rm 491, Baltimore, Maryland 21287, USA. Phone: 410.502.6244; Email: snimmag1@jhmi.edu.
Authorship note: DK and AL contributed equally to this work.
Find articles by Allaj, V. in: JCI | PubMed | Google Scholar
1The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Center for Translational Medicine, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
3Innovation Center for Biomedical Informatics, Georgetown University, Washington, DC, USA.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
6The Sidney Kimmel Comprehensive Cancer Center and the Bloomberg–Kimmel Institute for Cancer Immunotherapy, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
7Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
8Division of Clinical Pharmacology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Sridhar Nimmagadda, Johns Hopkins Medical Institutions, 1550 Orleans Street, CRB II, Rm 491, Baltimore, Maryland 21287, USA. Phone: 410.502.6244; Email: snimmag1@jhmi.edu.
Authorship note: DK and AL contributed equally to this work.
Find articles by
Sysa-Shah, P.
in:
JCI
|
PubMed
|
Google Scholar
|

1The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Center for Translational Medicine, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
3Innovation Center for Biomedical Informatics, Georgetown University, Washington, DC, USA.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
6The Sidney Kimmel Comprehensive Cancer Center and the Bloomberg–Kimmel Institute for Cancer Immunotherapy, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
7Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
8Division of Clinical Pharmacology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Sridhar Nimmagadda, Johns Hopkins Medical Institutions, 1550 Orleans Street, CRB II, Rm 491, Baltimore, Maryland 21287, USA. Phone: 410.502.6244; Email: snimmag1@jhmi.edu.
Authorship note: DK and AL contributed equally to this work.
Find articles by
Wharram, B.
in:
JCI
|
PubMed
|
Google Scholar
|

1The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Center for Translational Medicine, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
3Innovation Center for Biomedical Informatics, Georgetown University, Washington, DC, USA.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
6The Sidney Kimmel Comprehensive Cancer Center and the Bloomberg–Kimmel Institute for Cancer Immunotherapy, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
7Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
8Division of Clinical Pharmacology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Sridhar Nimmagadda, Johns Hopkins Medical Institutions, 1550 Orleans Street, CRB II, Rm 491, Baltimore, Maryland 21287, USA. Phone: 410.502.6244; Email: snimmag1@jhmi.edu.
Authorship note: DK and AL contributed equally to this work.
Find articles by Lesniak, W. in: JCI | PubMed | Google Scholar
1The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Center for Translational Medicine, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
3Innovation Center for Biomedical Informatics, Georgetown University, Washington, DC, USA.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
6The Sidney Kimmel Comprehensive Cancer Center and the Bloomberg–Kimmel Institute for Cancer Immunotherapy, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
7Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
8Division of Clinical Pharmacology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Sridhar Nimmagadda, Johns Hopkins Medical Institutions, 1550 Orleans Street, CRB II, Rm 491, Baltimore, Maryland 21287, USA. Phone: 410.502.6244; Email: snimmag1@jhmi.edu.
Authorship note: DK and AL contributed equally to this work.
Find articles by Tully, E. in: JCI | PubMed | Google Scholar
1The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Center for Translational Medicine, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
3Innovation Center for Biomedical Informatics, Georgetown University, Washington, DC, USA.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
6The Sidney Kimmel Comprehensive Cancer Center and the Bloomberg–Kimmel Institute for Cancer Immunotherapy, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
7Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
8Division of Clinical Pharmacology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Sridhar Nimmagadda, Johns Hopkins Medical Institutions, 1550 Orleans Street, CRB II, Rm 491, Baltimore, Maryland 21287, USA. Phone: 410.502.6244; Email: snimmag1@jhmi.edu.
Authorship note: DK and AL contributed equally to this work.
Find articles by Gabrielson, E. in: JCI | PubMed | Google Scholar
1The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Center for Translational Medicine, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
3Innovation Center for Biomedical Informatics, Georgetown University, Washington, DC, USA.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
6The Sidney Kimmel Comprehensive Cancer Center and the Bloomberg–Kimmel Institute for Cancer Immunotherapy, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
7Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
8Division of Clinical Pharmacology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Sridhar Nimmagadda, Johns Hopkins Medical Institutions, 1550 Orleans Street, CRB II, Rm 491, Baltimore, Maryland 21287, USA. Phone: 410.502.6244; Email: snimmag1@jhmi.edu.
Authorship note: DK and AL contributed equally to this work.
Find articles by Jaffee, E. in: JCI | PubMed | Google Scholar
1The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Center for Translational Medicine, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
3Innovation Center for Biomedical Informatics, Georgetown University, Washington, DC, USA.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
6The Sidney Kimmel Comprehensive Cancer Center and the Bloomberg–Kimmel Institute for Cancer Immunotherapy, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
7Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
8Division of Clinical Pharmacology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Sridhar Nimmagadda, Johns Hopkins Medical Institutions, 1550 Orleans Street, CRB II, Rm 491, Baltimore, Maryland 21287, USA. Phone: 410.502.6244; Email: snimmag1@jhmi.edu.
Authorship note: DK and AL contributed equally to this work.
Find articles by
Poirier, J.
in:
JCI
|
PubMed
|
Google Scholar
|

1The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Center for Translational Medicine, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
3Innovation Center for Biomedical Informatics, Georgetown University, Washington, DC, USA.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
6The Sidney Kimmel Comprehensive Cancer Center and the Bloomberg–Kimmel Institute for Cancer Immunotherapy, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
7Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
8Division of Clinical Pharmacology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Sridhar Nimmagadda, Johns Hopkins Medical Institutions, 1550 Orleans Street, CRB II, Rm 491, Baltimore, Maryland 21287, USA. Phone: 410.502.6244; Email: snimmag1@jhmi.edu.
Authorship note: DK and AL contributed equally to this work.
Find articles by
Rudin, C.
in:
JCI
|
PubMed
|
Google Scholar
|

1The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Center for Translational Medicine, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
3Innovation Center for Biomedical Informatics, Georgetown University, Washington, DC, USA.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
6The Sidney Kimmel Comprehensive Cancer Center and the Bloomberg–Kimmel Institute for Cancer Immunotherapy, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
7Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
8Division of Clinical Pharmacology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Sridhar Nimmagadda, Johns Hopkins Medical Institutions, 1550 Orleans Street, CRB II, Rm 491, Baltimore, Maryland 21287, USA. Phone: 410.502.6244; Email: snimmag1@jhmi.edu.
Authorship note: DK and AL contributed equally to this work.
Find articles by
Gobburu, J.
in:
JCI
|
PubMed
|
Google Scholar
|

1The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Center for Translational Medicine, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
3Innovation Center for Biomedical Informatics, Georgetown University, Washington, DC, USA.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
6The Sidney Kimmel Comprehensive Cancer Center and the Bloomberg–Kimmel Institute for Cancer Immunotherapy, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
7Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
8Division of Clinical Pharmacology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Sridhar Nimmagadda, Johns Hopkins Medical Institutions, 1550 Orleans Street, CRB II, Rm 491, Baltimore, Maryland 21287, USA. Phone: 410.502.6244; Email: snimmag1@jhmi.edu.
Authorship note: DK and AL contributed equally to this work.
Find articles by Pomper, M. in: JCI | PubMed | Google Scholar
1The Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
2Center for Translational Medicine, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
3Innovation Center for Biomedical Informatics, Georgetown University, Washington, DC, USA.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
6The Sidney Kimmel Comprehensive Cancer Center and the Bloomberg–Kimmel Institute for Cancer Immunotherapy, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
7Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
8Division of Clinical Pharmacology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Address correspondence to: Sridhar Nimmagadda, Johns Hopkins Medical Institutions, 1550 Orleans Street, CRB II, Rm 491, Baltimore, Maryland 21287, USA. Phone: 410.502.6244; Email: snimmag1@jhmi.edu.
Authorship note: DK and AL contributed equally to this work.
Find articles by
Nimmagadda, S.
in:
JCI
|
PubMed
|
Google Scholar
|

Authorship note: DK and AL contributed equally to this work.
Published November 20, 2018 - More info
J Clin Invest. 2019;129(2):616–630. https://doi.org/10.1172/JCI122216.
Copyright © 2019, American Society for Clinical Investigation
Received: June 5, 2018; Accepted: November 13, 2018
-
Abstract
Immune checkpoint therapies have shown tremendous promise in cancer therapy. However, tools to assess their target engagement, and hence the ability to predict their efficacy, have been lacking. Here, we show that target engagement and tumor-residence kinetics of antibody therapeutics targeting programmed death ligand-1 (PD-L1) can be quantified noninvasively. In computational docking studies, we observed that PD-L1–targeted monoclonal antibodies (atezolizumab, avelumab, and durvalumab) and a high-affinity PD-L1–binding peptide, WL12, have common interaction sites on PD-L1. Using the peptide radiotracer [64Cu]WL12 in vivo, we employed positron emission tomography (PET) imaging and biodistribution studies in multiple xenograft models and demonstrated that variable PD-L1 expression and its saturation by atezolizumab, avelumab, and durvalumab can be quantified independently of biophysical properties and pharmacokinetics of antibodies. Next, we used [64Cu]WL12 to evaluate the impact of time and dose on the unoccupied fraction of tumor PD-L1 during treatment. These quantitative measures enabled, by mathematical modeling, prediction of antibody doses needed to achieve therapeutically effective occupancy (defined as >90%). Thus, we show that peptide-based PET is a promising tool for optimizing dose and therapeutic regimens employing PD-L1 checkpoint antibodies, and can be used for improving therapeutic efficacy.
-
Introduction
The success of cancer drug therapy depends on a number of factors within the tumor including drug access, completeness of drug-target engagement, and targeting cancer cell heterogeneity. Target engagement in particular, which includes timing and extent of the drug-target interaction, is poorly understood for the majority of anticancer therapeutics (1–3). Existing technologies such as mass spectrometry (4), cellular thermal shift assay (5), or fluorescence anisotropy imaging (6) assist in the characterization of drug activity in cells and tissues. However, they have limited application in quantifying completeness of drug-target engagement within and across multiple lesions simultaneously, and in real time. Such real-time measurements are important to capture the dynamic changes in tumor target expression, which impacts success as in the case of immune checkpoint therapy (7–9). Noninvasive imaging technologies such as positron emission tomography (PET) provide a precise means to quantify drug-target engagement in all lesions concurrently and can be used to address some of those challenges (10). PET is used to quantify target engagement and enable dose-finding of some small-molecule therapeutics developed for central nervous system disorders, but only rarely for those developed for oncology and for biologics (11). Specifically, PET has not been used to define the drug-target engagement of immune checkpoint therapeutics. Given the complex tumor and tissue responses associated with immune checkpoint therapy, quantification of target expression and completeness of drug-target engagement, in principle, could be integrated into treatment protocols to improve efficacy, avoid off-target effects, and enable enrollment of patients most likely to respond.
Nearly 70% of patients receiving immune checkpoint therapeutics do not respond (12). To date, most research into immune checkpoint resistance has focused on genetic and immunological factors such as tumor mutational burden, T cell clonality, programmed death ligand-1 (PD-L1, also known as B7-H1) expression, and CD8-positive cells in the tumors (13–15). However, due to lack of suitable technology, the relevance of dose to the degree and duration of target engagement within tumors remains unknown for immune checkpoint therapeutics. Understanding these dynamic properties would contribute to a more complete understanding of the targeted intervention and ensure optimum immune response.
PD-L1–targeted therapeutics form the backbone of immune checkpoint therapy today, and methods for detecting and quantifying PD-L1 are critical for guiding such therapy (16). Three PD-L1 therapeutic mAbs (atezolizumab [AtzMab], avelumab [AveMab], and durvalumab [DurMab]) have received FDA approval; however, the extent of PD-L1 engagement by these mAbs at the tumor remains unknown. That is because intratumoral concentrations of antibodies are influenced by multiple factors including dynamic changes in PD-L1 expression, parameters intrinsic to tumors such as vascularization and interstitial pressure, and extrinsic parameters related to their complex pharmacokinetics influenced by drug-target-complex turnover (17). Peripheral T cell–derived target-engagement data are often used for dose-finding of PD-L1 therapeutics, but it may not truly reflect target engagement at the tumor. For example, for the antibody BMS-936559, a PD-L1 occupancy of 64%–70% has been reported in T cells for doses ranging from 0.1 to 10 mg/kg; however, occupancy and accessible PD-L1 levels in the tumor remain unknown (18). Radiolabeled antibodies can provide information about tumor accumulation at a given dose but have not been used to calculate the extent of PD-L1–drug engagement in patients, in part due to their prolonged circulation time (19). Such findings underscore the need for quantitative measurement of PD-L1 target levels and their engagement by therapeutic antibodies at the tumor, accounting for antibody kinetics, at all identifiable lesions.
We previously developed a high-affinity PD-L1–specific peptide radiolabeled with 64Cu, [64Cu]WL12, which generates high-contrast images within 120 minutes of radiotracer administration (20). Herein we evaluate [64Cu]WL12 PET to quantify dynamic changes in tumor PD-L1 expression and the potential for target engagement of FDA-approved PD-L1 therapeutics. Our results demonstrate that [64Cu]WL12 quantified PD-L1 engagement at the tumor independently of antibody biophysical characteristics and in vivo kinetics. We hypothesize that [64Cu]WL12 peptide-based PET will enable measurement of the PD-L1 engagement by any therapeutic antibody at the tumor to assist in improving outcomes in patients undergoing immunotherapy. These findings establish what we believe is a novel approach that can be generalized to study other therapeutic antibody interactions with their tumor target.
-
Results
Structural analysis of PD-L1 interactions with WL12 and PD-L1 mAbs. WL12 is a 14–amino acid circular peptide that binds PD-L1 with high affinity and disrupts interactions between PD-L1 and programmed death-1 receptor (PD-1) (IC50 20 nM). Our earlier molecular modeling analysis indicated an overlap in the interaction surface of PD-L1/WL12 and PD-L1/PD-1 complexes as the reason for the high affinity (20), which led us to hypothesize that the WL12 interaction surface on PD-L1 also overlaps with that of clinically available antibody therapeutics designed to inhibit PD-L1/PD-1 interactions. To test our hypothesis we compared the predicted binding conformations of WL12 with those of reported PD-L1 antibodies. The overlap of amino acid contacts between PD-L1 and PD-L1 antibodies, a PD-L1 nanobody (KN035), as well as PD-1 and WL12, revealed a common binding domain composed of PD-L1 residues Y56, E58, A113, M115, and Y123. As shown in the visualization of the PD-L1 molecular surface (Figure 1 and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI122216DS1), a singular overlapping region (cyan) forms a deep binding pocket, and acts as an anchor point for interactions with all of the agents investigated. The presence of a shared binding interface (cyan) is a strong indication that WL12 (green) competes with both the natural ligand (PD-1, purple) and antibodies. Further supporting our PD-L1/WL12 structural analysis, the shared binding interface has been observed with other macrocyclic peptide inhibitors targeted at disrupting PD-L1/PD-1 interactions (21). In terms of surface area, AtzMab (cyan and red) interacts with more of the PD-L1 surface than the other agents, with loops from the AtzMab generating molecular contacts with residues on all sides of the shared binding interface. Also, AtzMab overlaps with the interaction surfaces of PD-1 (cyan and purple), WL12 (cyan and green), AveMab (cyan and orange), and DurMab (cyan and blue). On the other hand, DurMab and AveMab show less overlap with the PD-1 interaction surface on PD-L1, further supporting the hypothesis that a shared binding interface pocket must be a core in the functional interaction for all the investigated agents. Those observations provide evidence that WL12 could be useful to detect and quantify antibody binding to the target.
 Figure 1
Figure 1WL12 binding interface on PD-L1 overlaps with PD-1 and PD-L1 therapeutics. (A) WL12 binding mode to PD-L1 (green and cyan) overlaps those of PD-1 (purple and cyan), AtzMab (red and cyan), AveMab (orange and cyan), and DurMab (blue and cyan). Noninteracting residues are shown in gray. The variety of contacts encompassing the shared binding region (cyan) illustrate the diverse binding mechanisms of different therapeutic antibodies.
In vitro validation of PD-L1 interactions with WL12 and PD-L1 mAbs. To validate the observations from structural analysis, we prepared Cy5-labeled AtzMab, AveMab, and DurMab analogs through conjugation of the fluorescent Cy5 N-hydroxysuccinimide (NHS) ester to each mAb. Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometric analyses of the purified conjugates showed an average of 1 to 2 Cy5 molecules per mAb (Supplemental Figure 2A). All 3 Cy5-mAb conjugates detected variable levels of PD-L1 expression in cell lines, with minor differences in mean fluorescence intensities (MFIs), and consistent with routinely used commercial PD-L1 detection antibodies (Supplemental Figure 2B). To assess the specificity of WL12 for inhibiting binding of antibodies to PD-L1, we performed competitive inhibition assays in CHO cells constitutively expressing PD-L1 (hPD-L1) and in MDAMB231 triple-negative breast cancer (TNBC) cells, which naturally express PD-L1 (22). We observed that WL12 dose-dependently inhibits Cy5-mAb binding to PD-L1, with IC50 values of 2 to 5 nM (Figure 2, A and B, and Table 1). We also validated those observations in HCC827 and H226 non–small cell lung cancer (NSCLC) cells, each of which naturally expresses moderate levels of PD-L1. HCC827 and H226 cells were incubated with Cy5-AtzMab, -AveMab, and -DurMab in the presence of a single concentration of WL12 (5 nM) and analyzed by flow cytometry for unoccupied–PD-L1 levels. Indeed, a significant reduction (P < 0.001) in Cy5-mAb binding indicating low unoccupied–PD-L1 levels was detected in the presence of WL12. We obtained further confirmation of the specificity of the WL12/PD-L1 interaction through the absence of a change in bound fluorescence when MDX1338, a CXCR4-specific (control) antibody, was used. Data in multiple cell lines with variable PD-L1 expression levels established the capacity of WL12 to disrupt antibody-PD-L1 interactions (Supplemental Figure 2, C and D). We next performed a bioluminescence-based functional assay to demonstrate the ability and potency of WL12 peptide to inhibit PD-1/PD-L1 interactions compared to PD-L1 antibodies (Supplemental Figure 2E). An in vitro functional assay showed that WL12 peptide is 10-fold less potent than AtzMab in inhibiting PD-1/PD-L1 interactions and therefore unlikely to interfere with an ongoing antibody therapy.
 Figure 2
Figure 2WL12 inhibits interaction between PD-1 and PD-L1 therapeutics in vitro. (A) Schematic representation of the assay. (B) WL12 inhibits Cy5-conjugated AtzMab, AveMab, and DurMab binding to PD-L1, as demonstrated through competitive inhibition and corresponding IC50 values. Mean fluorescence intensities (MFIs) were determined by flow cytometry. (C) Schematic representation of the assay. (D) [64Cu]WL12 binding to PD-L1–positive HCC827, H226, hPD-L1, and MDAMB231 cells is inhibited in the presence of 60 nM AtzMab, AveMab, and DurMab, compared with PBS control. [64Cu]WL12 binding in PD-L1–negative CHO and SUM149 cells is also shown. ****P < 0.0001; NS, not significant, by 1-way ANOVA and Dunnett’s multiple comparisons test in D.
Mutation in PD-L1 could impact binding of PD-L1 imaging agents and therapeutics. Analysis of cBioportal and CCLE databases indicated mutations in PD-L1 in a small percentage of tumors and cell lines (Supplemental Figure 2, F–H). Uptake studies of [64Cu]WL12 in HCC1569 cells with an M115T mutation that is at the interface of PD-L1/WL12 showed significant increase in [64Cu]WL12 uptake in IFN-γ–treated cells compared with untreated cells (Supplemental Figure 2I). These observations warrant further investigation into PD-L1 variants and their relevance to binding of PD-L1 inhibitors.
Next we sought to characterize the potential of the WL12 peptide–derived radiotracer to measure target engagement by PD-L1 antibodies. Previously, we showed that [64Cu]WL12 bound PD-L1 with high selectivity in vitro in hPD-L1 and CHO cells. However, such selectivity has not been validated in human cancer cell lines with variable endogenous expression (20). Accordingly, we selected multiple cell lines with high endogenous PD-L1 expression (HCC827, H226, MDAMB231, and hPD-L1). Cells were incubated with [64Cu]WL12 at 4°C for 30 minutes, washed, and cell-bound activity was measured. We observed high expression-dependent uptake of [64Cu]WL12 in those cells and uptake in PD-L1–positive cells reflected variable levels of surface PD-L1 expression observed by flow cytometry (Supplemental Figure 2B) in the order hPD-L1 > MDAMB231 > HCC827 > H226. PD-L1 specificity of [64Cu]WL12 uptake was low in PD-L1–negative cells (SUM149 and CHO, P < 0.0001). We observed significant blockade of [64Cu]WL12 uptake in all PD-L1–positive cells treated with 60 nM mAb compared with PBS-treated controls (P < 0.0001), indicating binding specificity of the radiotracer (Figure 2, C and D). The in vitro results suggest that [64Cu]WL12 could be used to quantify PD-L1/antibody interactions and unoccupied–PD-L1 levels in tumors.
In vivo quantification of tumor PD-L1 engagement by AtzMab. NSCLC xenograft models were used to evaluate in vivo PD-L1 engagement by the therapeutic mAbs in a noninvasive manner. We selected those models because nearly 50% of NSCLCs are PD-L1 positive and PD-L1 IHC is used as a predictive biomarker in patients with NSCLC undergoing immune checkpoint therapy (23).
We used NOD SCID γ (NSG) mice bearing H226 and HCC827 cell–derived xenografts that exhibit low and moderate PD-L1 expression, respectively, and treated them with a single dose of AtzMab (20 mg/kg) administered intravenously as a bolus, 24 hours before [64Cu]WL12 injection. PET images acquired 2 hours after [64Cu]WL12 injection and relatively 26 hours after AtzMab injection showed higher accumulation of [64Cu]WL12 in HCC827 tumors compared with H226. There was a clear reduction in the accumulation of radioactivity in tumors of AtzMab-treated mice, indicating reduced levels of available PD-L1 interaction sites compared with saline-treated controls (Figure 3, A and B). The PET imaging results were further confirmed by ex vivo measurements of biodistribution (Figure 3, D and E, and Supplemental Figure 3, A and B), which showed significant reductions in [64Cu]WL12 accumulation (as percentage of injected dose per gram of tissue [%ID/g]) in AtzMab-treated mice compared with saline controls: 34% (P < 0.0001) in H226 and 47% (P < 0.001) in HCC827 xenografts. Radiotracer uptake was corroborated by high PD-L1 staining intensity observed in HCC827 xenografts compared with H227 tumors (Figure 3, C and F). The results demonstrate that [64Cu]WL12 can be used to assess, qualitatively and quantitatively, in vivo targeting of PD-L1 at the tumor by AtzMab.
![PD-L1 engagement by anti–PD-L1 mAbs is quantified at the tumor using [64Cu]WL12 in xenografts with variable PD-L1 expression. PD-L1 engagement by anti–PD-L1 mAbs is quantified at the tumor using [64Cu]](//dm5migu4zj3pb.cloudfront.net/manuscripts/122000/122216/small/JCI122216.f3.gif) Figure 3
Figure 3PD-L1 engagement by anti–PD-L1 mAbs is quantified at the tumor using [64Cu]WL12 in xenografts with variable PD-L1 expression. (A–H) Reduced uptake of [64Cu]WL12 in H226 (A and B), HCC827 (C and D), and contralateral hPD-L1 and PD-L1–negative CHO (hPD-L1/CHO) (G and H) xenografts in mice treated with 20 mg/kg AtzMab 24 hours prior to radiotracer injection, compared with saline-treated controls. Whole-body, volume-rendered [64Cu]WL12 PET-CT images (A, D, and G) and ex vivo biodistribution (B, E, and H) at 2 hours after [64Cu]WL12 injection (n = 8–12/group). (C, F, and I) IHC staining for PD-L1 is shown from the corresponding tumor. Scale bars: 100 μm. Box-and-whisker graphs showing minimum to maximum and all data points, with the horizontal line representing the median. ****P < 0.0001; ***P < 0.001; NS, not significant, by unpaired Student’s t test in B, E, and H.
Expression of PD-L1 varies highly between and across lesions in a given patient and tumor type such that different drug concentrations may be needed to establish a similar extent of target engagement and to reduce immune-related toxicities. To measure the effect of a single dose of AtzMab on targeting different PD-L1 levels in tumors, we performed PET and biodistribution studies in tumors derived from the hPD-L1 cells, which have 4- to 10-fold higher PD-L1 expression than HCC827 and H226 cells (Supplemental Figure 2B). The mice bearing contralateral hPD-L1 and CHO tumors (hPD-L1/CHO) were treated with AtzMab (20 mg/kg for 24 hours) and changes in tumor uptake of [64Cu]WL12 were evaluated by PET. We observed high uptake of [64Cu]WL12 in hPD-L1 tumors (Figure 3G, right flank) compared with CHO tumors (left flank). Confirming AtzMab specific PD-L1 blockade, a significant reduction in [64Cu]WL12 uptake was observed in hPD-L1 tumors in AtzMab-treated mice, compared with saline-treated controls (Figure 3G). Validating PET imaging data and revealing significant tumor PD-L1 engagement by AtzMab, biodistribution studies showed a 77% reduction in [64Cu]WL12 uptake in the AtzMab-treated hPD-L1 tumors compared with saline controls (Figure 3H and Supplemental Figure 3C) (P < 0.0001). In contrast, we observed only a low level of [64Cu]WL12 uptake in PD-L1–negative control CHO tumors. Also, [64Cu]WL12 uptake in hPD-L1 tumors treated with AtzMab was not significantly different from that of CHO tumors, suggesting that treatment with a single 20 mg/kg dose of AtzMab led to near saturation of PD-L1 in those tumors. Those observations were confirmed by strong and weak immunoreactivity observed in hPD-L1 and CHO tumors, respectively (Figure 3I). Taken together, results from in vivo studies demonstrate that [64Cu]WL12 PET can detect graded levels of PD-L1 expression in tumors, and can be used to quantify a wide range of PD-L1 engagement by a single 20 mg/kg dose of AtzMab in the tumors.
Quantifying dynamic changes in PD-L1 expression. PD-L1 is upregulated in response to various cytokines, importantly IFN-γ, which contributes to dynamic and spatiotemporal heterogeneity in PD-L1 expression (24, 25). However, those rapid changes have not been quantified in vivo in real time. We sought to evaluate [64Cu]WL12 for its ability to quantify inducible PD-L1 expression within tumors in vivo (Figure 4), and to determine whether the blockade of such upregulated PD-L1 by AtzMab treatment could be monitored by [64Cu]WL12 PET.
![Dynamic changes in tumor PD-L1 expression and its engagement by AtzMab detected using [64Cu]WL12. Dynamic changes in tumor PD-L1 expression and its engagement by AtzMab dete](//dm5migu4zj3pb.cloudfront.net/manuscripts/122000/122216/small/JCI122216.f4.gif) Figure 4
Figure 4Dynamic changes in tumor PD-L1 expression and its engagement by AtzMab detected using [64Cu]WL12. (A) Flow cytometry histogram showing increased PD-L1 cell surface expression in A549-iPDL1 cells treated with doxycycline for 6 hours and 72 hours. (B) WL12 inhibits (5 nM) binding of Cy5-conjugated AtzMab, AveMab, and DurMab (2 nM) to A549-iPDL1 cells treated with doxycycline for 72 hours. (C) [64Cu]WL12 binding to A549-iPDL1 cells (72-hour doxycycline) is significantly reduced in the presence of 60 nM AtzMab, compared with controls. (D and E) [64Cu]WL12 uptake in A549-iPDL1 xenografts is significantly lower in mice receiving intravenous AtzMab 24 hours prior to radiotracer injection, compared with saline controls and similar to parent A549 xenografts. Volume-rendered whole-body PET-CT images (D), and ex vivo quantification (E) at 2 hours after [64Cu]WL12 injection (n = 10/group). (F) IHC staining for PD-L1 of the corresponding tumors. Scale bars: 100 μm. Box-and-whisker graphs showing minimum to maximum and all data points, with the horizontal line representing the median. ****P < 0.0001; NS, not significant, by 1-way ANOVA and Sidak’s multiple comparisons test in C and E.
We first generated an A549 NSCLC cell line with doxycycline-inducible PD-L1 expression (A549-iPDL1). A549 is a KRASG12S human lung adenocarcinoma cell line that expresses low PD-L1 at baseline. It has been transduced with PD-L1 in the all-in-one lentivirus pINDUCER20 vector (26), selected with G418, confirmed for PD-L1 induction by flow cytometry (Figure 4A), and used for in vitro and in vivo studies. Validating the PD-L1 specificity of WL12, binding of Cy5–PD-L1 mAbs to doxycycline-treated A549-iPDL1 cells was blocked in the presence of WL12 (Figure 4B). Next, A549-iPDL1 cells were treated with doxycycline for 72 hours, incubated with radiotracer for 30 minutes, and cell-associated radioactivity was measured. Doxycycline-treated A549-iPDL1 cells showed increased surface PD-L1 expression, and exhibited a 5.5-fold higher uptake of [64Cu]WL12, compared with untreated cells (P < 0.0001). We observed no significant differences in uptake between untreated A549-iPDL1 and parental A549 cells, confirming that [64Cu]WL12 binding was specific to induced PD-L1 expression. Furthermore, [64Cu]WL12 binding to doxycycline-treated A549-iPDL1 cells significantly decreased (65%, P < 0.0001) in the presence of 60 nM AtzMab, AveMab, and DurMab or nonradioactive WL12 (Figure 4C).
We next sought to validate those in vitro observations in vivo. NSG mice bearing A549-iPDL1 (Figure 4D, red circles) and A549 tumors (black circles) and treated with doxycycline for 72 hours were used. [64Cu]WL12 PET images and biodistribution studies of those mice showed high radioactivity uptake in A549-iPDL1 tumors. [64Cu]WL12 in A549-iPDL1 tumors was 65% higher than in the A549 control tumors (P > 0.0001). Following AtzMab treatment, [64Cu]WL12 uptake in A549-iPDL1 tumors was decreased more than 75%, with %ID/g values similar to control A549 tumors (Figure 4, D and E, and Supplemental Figure 4). IHC analysis of the tumors demonstrated an intense PD-L1 signal in A549-iPDL1 but not in A549 tumors, confirming imaging and biodistribution results (Figure 4F). Overall, the results demonstrate the potential of [64Cu]WL12 to detect dynamic changes in PD-L1 expression levels, and its blockade by AtzMab.
Quantifying target engagement in the tumors by different PD-L1 antibodies. We (22) and others (27–31) developed radiolabeled anti–PD-L1 antibodies and demonstrated their potential to assess PD-L1 expression in human tumor xenografts and syngeneic murine tumor models noninvasively. Observations made using radiolabeled antibodies, however, are highly specific to the antibody under investigation. In addition to the 3 FDA-approved PD-L1 mAbs, several others are in clinical development (32). A new approach is needed that (a) accounts for target engagement of PD-L1 antibodies, which (b) takes into account the properties of antibodies, and (c) is applicable to all PD-L1–targeted antibodies. Based on our in silico observations, we hypothesized that [64Cu]WL12 PET could be used to detect target engagement of any PD-L1 antibody that inhibits PD-L1/PD-1 interactions.
To test our hypothesis, we investigated the ability of [64Cu]WL12 PET to quantify PD-L1 engagement at the tumor by each of the 3 FDA-approved anti–PD-L1 mAbs. Moreover, those 3 mAbs exhibit distinct pharmacokinetics: AtzMab (isotype, IgG1κ; KD, 0.4 nM; t1/2, 27 days), AveMab (IgG1λ, 0.7 nM, 6.1 days), and DurMab (IgG1κ, 0.022 nM, 18 days) (33). MDAMB231 tumor–bearing NSG mice were treated with a single dose of AtzMab (20 mg/kg), or AveMab (10 mg/kg), or DurMab (10 mg/kg) for 24 hours, and received [64Cu]WL12 for PET and biodistribution studies (Figure 5, A–D). [64Cu]WL12 uptake was low in the tumors in all mAb-treated mice, compared with saline controls, confirming low levels of unoccupied PD-L1 in the tumors due to PD-L1 engagement by the mAbs and radiotracer blockade. Ex vivo quantification of tumors validated those observations and demonstrated approximately 60% less uptake of [64Cu]WL12 in the tumors of mAb-treated mice (P < 0.0001) compared with saline controls (Figure 5E and Supplemental Figure 5). We have not observed significant differences in tumor PD-L1 blockade by different mAbs at the investigated dose and time, suggesting that all the mAbs demonstrate robust PD-L1 blockade at the tumor at early time points. IHC of saline controls demonstrated moderate to high PD-L1 intensity, placing these xenografts between highly PD-L1–positive hPD-L1 and weakly positive HCC827 and H226 tumors (Figure 5F). PD-L1 antibody treatment resulted in increased PD-L1 expression as an adaptive immune response to therapy in responders (34). Treatment of cell lines with PD-L1 mAbs had no effect on PD-L1 protein expression in the cell lines tested (Supplemental Figure 5B). The results demonstrate that tumor PD-L1 engagement by PD-L1 antibodies can be quantified by [64Cu]WL12 PET and independently of the plasma and tissue kinetics exhibited by a given PD-L1 antibody. Importantly, this approach could provide a method to evaluate biosimilar antibodies as they are developed to facilitate the time to approval and reduce clinical testing expense.
![Tumor PD-L1 engagement by 3 distinct PD-L1 therapeutic antibodies quantified using [64Cu]WL12. Tumor PD-L1 engagement by 3 distinct PD-L1 therapeutic antibodies quantifie](//dm5migu4zj3pb.cloudfront.net/manuscripts/122000/122216/small/JCI122216.f5.gif) Figure 5
Figure 5Tumor PD-L1 engagement by 3 distinct PD-L1 therapeutic antibodies quantified using [64Cu]WL12. (A–E) [64Cu]WL12 uptake in MDAMB231 xenografts is significantly reduced in mice receiving AtzMab (20 mg/kg), AveMab (10 mg/kg), or DurMab (10 mg/kg), 24 hours prior to radiotracer injection. Whole-body volume-rendered [64Cu]WL12 PET-CT images of saline- (A), AtzMab- (B), AveMab- (C), and DurMab-treated (D) mice, and ex vivo biodistribution (E) at 2 hours after [64Cu]WL12 injection (n = 6–9/group). (F) IHC staining for PD-L1 in the corresponding tumor. Scale bars: 100 μm. Box-and-whisker graphs showing minimum to maximum and all data points, with the horizontal line representing the median. ****P < 0.0001; NS, not significant, by 1-way ANOVA and Dunnett’s multiple comparisons test in E.
Effect of dose and time on PD-L1 occupancy at the tumor. Antibody kinetics at the tumor are complex and governed by both tumor-intrinsic and -extrinsic parameters (17). We (22) and others (29) recently discovered that factors other than PD-L1 expression can reduce accumulation of the PD-L1–targeted therapeutic AtzMab and its mouse chimera (PRO304397) within NSCLC, TNBC, and colon tumors. Furthermore, experiments in mice showed that at doses less than 1 mg/kg, the radioactivity measured from the systemically injected, radiolabeled PRO304397 was primarily associated with tumor vasculature, and showed minimal diffusion into tumor parenchyma in PD-L1–expressing tumor models (29). Those findings may be attributable to factors such as elevated intratumoral interstitial pressure, which impedes accumulation of antibodies in tumors, and estimated to be approximately 10%–30% of plasma levels (35–37). Such effects might also impede the access of large PD-L1–directed agents to targeted tumor cells and immune infiltrates. Occupancy measurements of PD-L1 therapeutics have been restricted to assessments made using peripheral blood mononuclear cells (PBMCs), in order to determine the dissociation constants of the drugs. However, PD-L1 occupancy measurements at the tumor have not yet been reported, and could provide a direct readout of therapeutic activity in the tumor.
To evaluate the effect of anti–PD-L1 therapeutic dose on tumor PD-L1 occupancy at the tumor, mice bearing MDAMB231 tumors were treated with single doses of AtzMab (0.06, 0.6, and 3.6 mg/kg body weight). Imaging and biodistribution studies were performed 2 hours after the injection of [64Cu]WL12 and relatively 26 hours after AtzMab administration. PET images of mice that received 0.06 mg/kg showed high [64Cu]WL12 uptake, indicating low PD-L1 occupancy by AtzMab at the tumor (Figure 6, A and B). At the higher doses (0.6 and 3.6 mg/kg), there was a proportional decrease in [64Cu]WL12 uptake in the tumors, indicating proportionally higher target engagement at the tumor by AtzMab. Corresponding biodistribution studies also demonstrated reduced unoccupied–PD-L1 levels in the tumors with increased dose, confirming that antibody dose–related changes in PD-L1 occupancy could be measured using [64Cu]WL12.
![Effect of dose and time on the unoccupied fraction of tumor PD-L1 following treatment with AtzMab quantified using [64Cu]WL12. Effect of dose and time on the unoccupied fraction of tumor PD-L1 following](//dm5migu4zj3pb.cloudfront.net/manuscripts/122000/122216/small/JCI122216.f6.gif) Figure 6
Figure 6Effect of dose and time on the unoccupied fraction of tumor PD-L1 following treatment with AtzMab quantified using [64Cu]WL12. (A and B) Dose-exposure relationship depicting the decrease in unoccupied PD-L1, in MDAMB231 tumors in mice, with increase in AtzMab dose (mg/kg). (A) Whole-body [64Cu]WL12 PET-CT images of MDAMB231 tumor–bearing mice receiving 0.06, 0.6, and 3.2 mg/kg AtzMab. (B) Ex vivo quantification of [64Cu]WL12 uptake in tumors of mice treated with escalating doses of AtzMab (0.06, 0.6, and 10 mg/kg). AtzMab was injected 24 hours prior to radiotracer injection. (C and D) AtzMab dose effect on tumor PD-L1 occupancy over time depicting an increase in unoccupied PD-L1 with 0.6 or 1 mg/kg dose of AtzMab, but not with 10 or 20 mg/kg AtzMab dose, recapitulating the nonlinear kinetics of mAb. Whole-body volume-rendered [64Cu]WL12 PET-CT images (C) and ex vivo biodistribution (D) at 2 hours after [64Cu]WL12 injection (n = 6–9/group). Box-and-whisker graphs showing minimum to maximum and all data points, with the horizontal line representing the median. *P < 0.05, ****P < 0.0001; NS, not significant, by unpaired Student’s t test in D.
Next, we sought to investigate the temporal changes in PD-L1 occupancy for a given AtzMab dose. Membrane-associated antigens such as PD-L1 can enhance antibody clearance through target-mediated endocytosis. Additionally, development of anti-therapeutic antibodies could significantly impact antibody serum and tumor kinetics (38). In support of those factors at play, earlier studies showed that AtzMab followed nonlinear kinetics below 0.6–1 mg/kg and linear kinetics above 1 mg/kg, and a tendency toward reduced serum concentration in patients that developed anti-therapeutic antibodies (39). All of those factors will influence unoccupied–PD-L1 levels and tumor exposure and response to therapy. The tools to measure the effect of those factors on the tumor exposure to the anti–PD-L1 therapeutics in real time have not been described.
To investigate the ability of [64Cu]WL12 PET to detect temporal changes in antibody kinetics at the tumor, MDAMB231 tumor–bearing NSG mice were injected with AtzMab doses that were described to be associated with nonlinear (0.6 and 1 mg/kg) and linear kinetics (10 and 20 mg/kg), and PET imaging and biodistribution studies were performed at 24 and 120 hours. At 24 hours, there was a significant reduction in [64Cu]WL12 uptake, also reflected in tumor %ID/g values, in all 4 dose groups, compared with saline-treated controls (Figure 6, C and D). At 120 hours, there was a significant increase in [64Cu]WL12 uptake in the 0.6 and 1 mg/kg dose groups compared with 24 hours. At 120 hours, [64Cu]WL12 uptake in the tumor was similar in 0.6 mg/kg–treated and saline control groups, suggesting the more rapid elimination of the drug from the tumor at the lowest dose. At the higher doses, however, [64Cu]WL12 uptake was significantly different from the saline control group, suggesting a slower elimination of the drug from the tumor, due to the saturable elimination or nonlinear pharmacokinetics of the drug at the higher doses. The temporal difference in [64Cu]WL12 uptake was not significant in the 10 or 20 mg/kg treatment groups, reflecting the linear pharmacokinetics or constant clearance of AtzMab at those doses. Collectively, the results show that in a mouse model both dose- and time-dependent changes in PD-L1 engagement can be quantitatively evaluated at the tumor.
Modeling dose-receptor occupancy of PD-L1 antibodies at the tumor. Pharmacokinetic-pharmacodynamic (PK/PD) modeling and simulation, linking the PK of the mAb to its receptor occupancy and tumor response, are routinely used as a modern approach for guiding starting dose selection from preclinical data, for first-in-human clinical trials, and for dose-range selection, in the later efficacy/safety clinical trials. Linking the receptor occupancy to the PK/PD model provides a mechanistic understanding of the antibody dose/concentration and tumor response relationship, and is valuable for efficient drug development and optimal dose selection (17, 40). PD-L1 expression in the tumors can be constitutive or induced, and its cell-surface stability could be modulated by oncogenic signaling and increased expression of positive regulators (7–9) that are difficult to account for based solely on in vitro assays. Those PK/PD mathematical models could be further refined using measurements of PD-L1 occupancy made at the tumor that encapsulate all the factors regulating PD-L1 levels and account for complex PD-L1 antibody kinetics.
To demonstrate the use of receptor occupancy measurements made at the tumor to define target-engagement characteristics and for dose finding, we measured unoccupied–PD-L1 levels in the tumors following AtzMab treatment. Mice bearing MDAMB231 tumors were injected with escalating doses of AtzMab from 0.009 to 24 mg/kg body weight. Twenty-four hours after AtzMab injections, biodistribution studies were performed 2 hours after the injection of [64Cu]WL12. Because of the large number of groups involved, we relied on ex vivo biodistribution studies. A developed inhibitory sigmoidal Emax model appropriately fitted and described the relationship between the dose of the AtzMab used in our experiment and the decrease in unoccupied PD-L1 at the tumor (Figure 7, A and B, and Supplemental Figure 6), detected using [64Cu]WL12 and expressed as %ID/g. The dose of the AtzMab responsible for 50% of the maximum PD-L1 engagement in the tumor (ID50) was estimated to be 0.43 mg/kg (Table 2). The ID90 and ID96, responsible for 90% and 96% of the maximum fractional decrease in unoccupied PD-L1 from baseline (Imax), corresponded to 0.87 mg/kg and 1.19 mg/kg, respectively. These dose levels are comparable to the dose of 1 mg/kg reported by Deng et al. for the chimeric anti–PD-L1 antibody PRO304397 and responsible for maximum PD-L1 saturation over 24 hours in BALB/c mice (29). Assuming a similar average volume of distribution at steady state (Vss) of 50 ml/kg for the anti–PD-L1 and the chimeric anti–PD-L1 antibody PRO304397 (29), the expected average plasma concentrations resulting from ID50, ID90, and ID96 were tentatively estimated to be 59 nM (8.6 μg/ml), 120 nM (17.4 μg/ml), and 164 nM (23.8 μg/ml), respectively. These results indicate the potential of using measurements made at the tumor, which reflect true target occupancy, for dose selection and optimization of future PD-L1 therapeutics.
![Effect of dose and time on tumor PD-L1 occupancy by AtzMab quantified using [64Cu]WL12. Effect of dose and time on tumor PD-L1 occupancy by AtzMab quantified using](//dm5migu4zj3pb.cloudfront.net/manuscripts/122000/122216/small/JCI122216.f7.gif) Figure 7
Figure 7Effect of dose and time on tumor PD-L1 occupancy by AtzMab quantified using [64Cu]WL12. (A and B) Dose-exposure relationship depicting the decrease in unoccupied PD-L1, in MDAMB231 tumors in mice, with increase in AtzMab dose (mg/kg). Ex vivo quantification of [64Cu]WL12 uptake at 2 hours in tumors of mice treated with escalating doses of AtzMab (0.0009 to 24 mg/kg) (n = 3–9/group). AtzMab was injected 24 hours prior to radiotracer injection (A). Percentage of unoccupied PD-L1 was calculated relative to the median unoccupied PD-L1 measured at 0 mg/kg (B). Blue open dots: measured unoccupied PD-L1 for each dose level in mice. Red dashed line: mean model-predicted dose-response relationship. (C) Schematic of PD-L1 PET for PD-L1 therapeutic development and evaluation. sPD-L1, soluble PD-L1; TMDD, target-mediated drug disposition.
 Table 2
Table 2Pharmacodynamic model parameters describing the relationship between anti–PD-L1 antibody dose and the decrease in unoccupied PD-L1 measured at the tumor site
-
Discussion
We have demonstrated the ability to measure PD-L1 occupancy using peptide-based PET according to a method that is agnostic to the choice of antibody. We were able to noninvasively quantify target engagement and pharmacokinetics of the PD-L1 mAbs at the tumor. The real-time measurements described encapsulate (a) the complex serum and tumor kinetics (or fate) of antibodies at the tumor site, (b) tumor-intrinsic parameters such as high interstitial pressure and poor vascularity that impede antibody accumulation, and (c) genetic and epigenetic mechanisms that regulate PD-L1 expression levels and turnover (Figure 7C). We further demonstrated the translational utility of our approach, by combining our experimentally derived, unoccupied–PD-L1 levels in the tumors with reported preclinical pharmacokinetics of AtzMab, to find a dose to achieve minimum efficacious occupancy levels at the tumor, as previously suggested (29).
Biologics such as antibodies pose unique challenges in predicting in vivo target engagement based on in vitro binding affinity (17). For small molecules, in vitro binding affinity measurements and target occupancy studies are routinely used for dose selection in central nervous system diseases and are predictive of pharmacological response (41). However, properties intrinsic to antibodies, such as valence, shape, size, and isoelectric point, make the pharmacokinetics of antibodies more complex and difficult to predict. Also, several parameters intrinsic to the tumor, such as antigen density and turnover, tumor burden, and tumor perfusion, limit intratumoral penetration of antibodies. Tumor and plasma concentrations of antibodies are further influenced by factors extrinsic to tumors such as dose, patient variability, cachexia, and development of anti-therapeutic antibodies (42). All of these factors affect tissue exposure of antibodies and ultimately efficacy. For example, pidilizumab, developed with investments of over $200 million, was later discovered not to bind PD-1, its intended target (43). Our approach accounts for target engagement of PD-L1 antibodies at the site of action and is applicable to all PD-L1 antibodies that inhibit PD-L1/PD-1 interactions.
The methods described here can be extended to evaluate the changes in unoccupied PD-L1 in all lesions in a patient concurrently, and used to further guide therapeutic decisions. Although IHC-based clinical tests are used to assess PD-L1 expression in tumors (34, 44, 45), PD-L1 IHC takes into account only a small fraction (0.1%) of a single lesion. Such an approach has significant limitations, because PD-L1 expression in the tumor microenvironment is spatially and temporally heterogeneous (46, 47). Also, tissue samples acquired by biopsy for testing are typically very limited, and may be needed for molecular profiling to identify targetable oncogenic mutations in other pathways that confer sensitivity or resistance to existing therapies (48). As a result of that heterogeneity, the sensitivity of a single small tissue sample can be low for detecting PD-L1 expression at scoring thresholds commonly used in clinical practice (49). Furthermore, immune therapy responses are delayed, complex, and abscopal in nature and it is impractical to perform multiple biopsies in multiple lesions for reliable representation of PD-L1 expression (50). Those issues are compounded in patients with metastatic disease, a population in which immune checkpoint therapeutics are extensively investigated.
New regulators of PD-L1 are emerging that control cell surface PD-L1 levels. The CKLF-like MARVEL transmembrane domain–containing proteins 4 and 6 (CMTM 4/6) (7, 8) and oncogenic proteins such as mutant RAS have been shown to increase cell surface PD-L1 protein lifetime or PD-L1 expression (9). Such PD-L1 dynamics are difficult to capture by IHC-based assessments and contribute to our limited success in advancing immunotherapies. Our studies with [64Cu]WL12 demonstrate that variable and dynamic changes in PD-L1 expression could be quantified during therapy and within the standard clinical work flow, yielding important clinical implications for patient selection and monitoring therapy. [64Cu]WL12 uptake in the tumors is highest at 60 minutes (14.9 %ID/g) and reduced by 15 to 12.6 %ID/g at 120 minutes (20). The image contrast is high at 120 minutes due to clearance from normal tissues. The fast clearance of the peptide suggests a washout from the tumor within 24 hours. Also, WL12 has lower binding affinity for PD-L1 than antibodies. Therefore, we do not expect [64Cu]WL12 to interfere with therapeutic efficacy of anti–PD-L1 antibodies, as it is given as a tracer dose (below μg/kg). Our [64Cu]WL12 PET measurements provide relative changes in target expression rather than true expression and such measurements acquired using PET are routinely used in the clinic (51). Other methods such as flow cytometry or autoradiography allow quantification of true expression; however, PET facilitates quantification of unoccupied–PD-L1 levels in real time and the measurements take all the PK/PD-induced changes in antibody concentration at the tumor into account that are difficult to capture using in vitro or ex vivo studies.
Until now, the tools to establish the relevance of dose to drug accumulation and target saturation, which is relevant to efficacy, have been unavailable for immune checkpoint therapeutics. Nearly 25% of all immune checkpoint therapy clinical trials target PD-L1, and nearly 70% of involved patients do not respond to treatment (12). What is unknown is whether the drug fully engages the target in vivo successfully, or whether the tumor is immunologically resistant. Another question is to what extent does tumor microenvironment (stroma and extracellular matrix [ECM]) contribute to immunological resistance by hindering therapeutic accumulation in some lesions. Tumors display an abnormal microenvironment and ECM that are shaped by the tumor location, and influence drug accumulation. Analysis of tissue from multiple disease sites from a patient with melanoma showed an upregulation of genes that define the ECM in nonresponding lesions (52). The role of the ECM in tumor resistance to antibody transport has been demonstrated previously, but has not been established in the context of immune checkpoint therapeutics (53). We have used subcutaneous lung cancer xenografts, which do not fully capture the microenvironment of the orthotopic tumors and could potentially influence antibody accumulation. Nevertheless, our PET imaging approach will enable measurements of total PD-L1 and its saturation by the antibodies at every identifiable tumor while accounting for changes in ECM, facilitating therapy optimization across multiple tumor types.
Radiolabeled antibodies such as AtzMab, supported by peripheral pharmacodynamics assessments and PK/PD modeling, are routinely used to predict the antibody dosing levels required to achieve the desired PD-L1 occupancy at the tumor (29). Those measurements of radiolabeled-antibody concentrations in the plasma and tumor, as well as the subsequent occupancy predictions derived through mathematical modeling, are often specific to a given antibody, limiting generalization of such observations to other PD-L1 antibodies (54). The absence of an effective, widely applicable method that accounts for complex antibody kinetics and provides real-time PD-L1 saturation/occupancy data impedes optimized dose selection and therapeutic development. Our in silico, in vitro, and in vivo data using [64Cu]WL12 PET demonstrate that PD-L1 saturation/occupancy can be noninvasively quantified at the tumor, a concept that can address this unmet need.
We provide a tool that can be used to assess antibody kinetics and target engagement potential for an ever-expanding array of PD-L1 therapeutic antibodies (32). Our data demonstrate that dynamic changes in PD-L1 expression in tumors, and PD-L1 saturation/occupancy by therapeutic antibodies, can be noninvasively measured independently of antibody selection. Imaging markers that directly reflect the levels and occupancy of PD-L1 have the potential to be more predictive, and form a bridge between dose, PD-L1 levels in the tumors, and therapeutic response. To the best of our knowledge our method provides the first noninvasive and translational tool to measure antibody target engagement at PD-L1, which is the necessary initial step toward finding safe, effective, and personalized doses for immunotherapy based on checkpoint inhibition. Importantly, this tool can be used to rapidly develop other tumor-targeted antibodies, significantly reducing time to approval and cost of development.
-
Methods
Study design. The goal of this study was to develop a noninvasive method to quantify the unoccupied fraction of PD-L1 and PD-L1 engagement by therapeutic antibodies. We performed a structural analysis of PD-L1, PD-1, and PD-L1 binding peptide ([64Cu]WL12) interactions and observed that [64Cu]WL12 could be used to evaluate the unoccupied fraction of PD-L1. We validated [64Cu]WL12 to quantify unoccupied–PD-L1 levels in vitro and in vivo using multiple cell lines and xenografts with graded levels of PD-L1 expression using 3 FDA-approved PD-L1 mAbs: AtzMab, AveMab, and DurMab. We then showed that [64Cu]WL12 can be used to evaluate the effect of dose of antibody and time on the unoccupied fraction of tumor PD-L1 following treatment, and used that data for the prediction of the dose of antibody needed to achieve therapeutically effective occupancy (defined as >90%) at the tumor by mathematical modeling.
Chemicals. WL12 was custom synthesized by CPC Scientific with greater than 95% purity. 2,2′,2′′-(10-(2,6-Dioxotetrahydro-2H-pyran-3-yl)-1,4,7,10-tetraazacyclododecane-1,4,7-triyl)triacetic acid (DOTAGA anhydride) was purchased from CheMatech Macrocycle Design Technologies (catalog C109). All other chemicals were purchased from Sigma-Aldrich or Thermo Fisher Scientific. We synthesized WL12-DOTAGA as previously described by us (20). [64Cu]CuCl2 was purchased from the University of Wisconsin (Madison, Wisconsin, USA) and Washington University (St. Louis, Missouri, USA).
Cell culture reagents and antibodies. All cell culture reagents were purchased from Invitrogen. The anti–human PD-L1 mAb AtzMab and anti-CXCR4 mAb ulocuplumab (55) were generated as described previously (56), and AveMab and DurMab were purchased from the Johns Hopkins Hospital pharmacy.
Docking studies. The structure files for PD-L1 bound to PD-1 (57), AveMab (58), AtzMab (59), DurMab (59), BMS936559 (60), and KN035 (61) were obtained from the Protein Data Bank (62), and compared to the predicted binding conformation of the cyclical peptide WL12 (20). The bound antibody and peptide structures were first aligned around the core of the PD-L1 interaction interface (residues 53–69), and the contact between the molecular surface of each antibody with PD-L1 was then identified using the Visual Molecular Dynamics software package (63).
Radiopharmaceutical preparation. 64CuCl2 was buffered with 100 mM sodium acetate solution (pH 5.5) and converted to 64Cu(OAc)2. For radiolabeling, approximately 20 μg of WL12-DOTAGA peptide conjugate (4.27 nmol), in 100 μl sodium acetate, was mixed with 3–5 mCi of 64Cu(OAc)2 and incubated at 65°C for 30 minutes. The resulting radiotracer was purified on a C-18 column (Luna, 5 μm, 10 × 250 mm; Phenomenex) using an elution gradient of 60% methanol (0.1% trifluoroacetic acid [TFA]) reaching 10% (with H2O, 0.1% TFA), over 30 minutes at a flow rate of 5 ml/min. HPLC was performed on a Varian ProStar system equipped with a radioactive single-channel radiation detector (model 105S; Bioscan) and a Varian ProStar UV absorbance detector set to 280 nm. [64Cu]WL12 was collected at 14.5 minutes, evaporated, formulated with saline containing 5% DMSO and 1 drop of Tween 20, and used for in vitro and in vivo evaluation. [64Cu]WL12 was obtained in 62.58% ± 1.9% yield, with a specific activity of 1.9 ± 0.11 mCi/μg.
Fluorescent labeling of AtzMab, AveMab, and DurMab. To conjugate the fluorophore to the therapeutic mAbs, we used Cy5 NHS ester (excitation/emission of 650/669). Briefly, 1 mg of mAb (6.5 nmol) in 400 μl of PBS was mixed with 4 μl of DMSO containing 40 μg of Cy5 NHS ester (65 nmol, GE, catalog 45-001-190). The reaction mixture was gently stirred for 2 hours at room temperature, followed by purification using Zeba spin desalting columns, 7K MWCO, 0.5 ml (Thermo Fisher Scientific, catalog 89882) pre-equilibrated in PBS. Analysis of the Cy5-mAbs, carried out using a Nanodrop 2000 UV-Vis spectrophotometer (Thermo Fisher Scientific), indicated a 1:2 mAb/Cy5 molar ratio.
MALDI-TOF analysis of antibodies. MALDI-TOF spectra of Cy5-conjugated and parent mAbs (AtzMab, AveMab, and DurMab) were obtained on a Voyager DE-STR MALDI-TOF available at the Johns Hopkins University Mass Spectrometry core facility. Briefly, mAb samples were equilibrated in water with 0.1% TFA using Amicon Ultra-15 centrifugal filter units (catalog UFC901008). Samples were mixed (1:2 dilution) with 10 mg/ml sinapinic acid (3,5-dimethoxy-4-hydroxycinnamic acid) matrix dissolved in 40% acetonitrile and 0.1% TFA. One microliter of sample was spotted in quadruplet on a MALDI plate (Applied Biosystems) and allowed to air dry, followed by spectra acquisition using optimized instrument settings. Data were analyzed using Applied Biosystems Data Explorer software version 4.8.
Cell culture. The A549, H226, HCC827, MDAMB231, and CHO cells were purchased from the American Type Culture Collection (ATCC). The CHO cells constitutively expressing PD-L1 (hPD-L1) were generated in our laboratory, and cultured as previously described (22). The SUM149 cells were obtained from Stephen Ethier (now at Medical University of South Carolina, Charleston, South Carolina, USA). All the cell lines were authenticated by STR profiling at the Johns Hopkins Genetic Resources Core Facility. The SUM149 cells were maintained in Ham’s F-12 medium with 5% FBS, 1% penicillin/streptomycin, 5 μg/ml insulin, and 0.5 μg/ml hydrocortisone. All other cell lines were cultured in ATCC-recommended media, in an incubator at 37°C in an atmosphere containing 5% CO2.
Inducible PD-L1 expression vector. To generate a doxycycline-inducible PD-L1 (B7-H1, CD274) lentiviral expression vector, we Gateway cloned the full-length cDNA from pENTR223-CD274 (ORFeome Collaboration clone ccsbBroadEn_03086; DNASU Plasmid Repository clone HsCD00508899) into pINDUCER20, a gift from Stephen Elledge (Harvard Medical School, Boston, Massachusetts, USA) (Addgene plasmid 44012), by LR reaction (26, 64). The reaction mixture was transformed into Stbl3 cells and selected on LB agar plates supplemented with 100 μg/ml carbenicillin. The purified recombinant plasmid (pINDUCER20-CD274) was analyzed via BsrGI restriction enzyme digestion, and the sequence confirmed by Sanger sequencing.
Lentiviral production. The day before transfection, 3.4 × 106 HEK-293T cells were plated in a 75-cm2 flask. The next day, the cells were transfected with lentiviral plasmids by polyethylinimine transfection, in a 3:2:1 ratio of pINDUCER20-CD274/psPAX2/pMD2.G. The lentiviral packaging plasmids pMD2.G (Addgene plasmid 12259) and psPAX2 (Addgene plasmid 12260) were gifts from Didier Trono (EPFL, Lausanne, Switzerland). At 24 hours after transfection, the media were changed to complete media. Seventy-two hours after transfection, the lentivirus was harvested, filtered through a 0.45-μm syringe filter, and stored at –80°C.
Lentiviral transduction. On day one, 1.5 × 105 A549 cells were plated in a 25-cm2 flask. On day 2, the cells were infected with 75 μl of lentiviral supernatant in 5 ml of complete media supplemented with 8 μg/ml polybrene. Twelve hours following lentiviral transduction, the media were removed and replaced with fresh complete media. To select for transduced cells, the media were replaced the following day with media containing G418 sulfate (Thermo Fisher Scientific, BP673) at a final concentration of 400 μg/ml. The PD-L1 expression in stable clones was analyzed by flow cytometry.
Detection of PD-L1 expression by flow cytometry and competitive inhibition assays. Cells were evaluated for PD-L1 surface expression by direct staining of 2 × 105 cells in 100 μl PBS with Cy5-conjugated PD-L1 mAbs, for 30 minutes at 4°C. Cells were then washed and analyzed for MFI by flow cytometry. Adherent cells were detached using enzyme-free cell dissociation buffer (Thermo Fisher Scientific).
Competitive inhibition assays were performed in 100 μl total volume in FACS buffer (1× PBS with 2 mM EDTA and 0.5% FBS). The WL12 peptide was dissolved in DMSO to generate a 412 μM stock to make 4× and 10× intermediate concentrations. WL12 was tested at 10 concentrations (0.1, 0.2, 0.3, 0.9, 2.7, 8.1, 15, 30, 60, and 100 nM). The WL12 stock (10 μl) was added to 2 × 105 hPD-L1 or MDAMB231 cells in 80 μl and incubated on ice for 30 minutes. Then, 10 μl of Cy5-conjugated mAb was added to the cells, to reach a final concentration of 2 nM for AtzMab, AveMab, and ulocuplumab, and 1 nM for DurMab, and incubated for 30 minutes at 4°C. Cells were then washed and analyzed for MFI by flow cytometry. The IC50 values were calculated based on normalized MFI values using GraphPad Prism 6 software. Data were acquired on an Accuri flow cytometer (Becton Dickinson) and at least 20,000 events were recorded.
In vitro binding assays with [64Cu]WL12. In vitro binding of [64Cu]WL12 to H226, HCC827, hPD-L1, CHO, MDAMB231, SUM149, A549, and A549-iPDL1 cells was determined by incubating 1 × 106 cells with approximately 0.1 μCi of [64Cu]WL12 in the presence or absence of 60 nM mAb for 30 minutes at 4°C. After incubation, cells were washed 3 times with ice-cold PBS containing 0.1% Tween 20, and counted on an automated gamma counter (1282 Compugamma CS, Pharmacia/LKBNuclear, Inc.). To demonstrate PD-L1–specific binding of [64Cu]WL12, blocking was performed with 1 μM nonradioactive WL12. All the cell radioactivity uptake studies were performed in quadruplicate for each cell line, and repeated 3 times.
In vivo studies. To establish xenografts, we used 5- to 6-week-old, male or female, nonobese, diabetic, severe-combined immunodeficient gamma (NSG) mice, obtained from the Johns Hopkins University Animal Resources Core.
Xenograft models. Mice were implanted with H226 (5 × 106, s.c.), HCC827 (5 × 106, s.c.), hPD-L1 (10 × 106, s.c.), CHO (10 × 106, s.c.), MDAMB231 (2 × 106, orthotopic), A549 (5 × 106, s.c.), or A549-iPDL (5 × 106, s.c.) cells in their upper flanks. If 2 cell lines were used, cells were inoculated in the opposite flanks. For treatment, imaging, or biodistribution experiments, mice with tumor volumes of 100–300 mm3 were used.
To induce PD-L1 expression in A549-iPDL1 tumors, mice were given doxycycline ad libitum (10 mg/ml doxycycline plus 1% sucrose) in drinking water 72 hours prior to [64Cu]WL12 injection.
For occupancy determination of AtzMab, AveMab, and DurMab at the tumor, xenograft-bearing mice (n = 9–12) were treated with a single intravenous bolus dose of either 20 mg/kg AtzMab, 10 mg/kg AveMab, or 10 mg/kg DurMab. Mice were used for imaging (n = 3–4) and biodistribution studies (n = 6–8) 24 hours after mAb treatment.
For determining dose effect on the unoccupied fraction of PD-L1 in the tumor, MDAMB231 tumor–bearing mice were treated with a single intravenous bolus escalating dose of AtzMab, and 24 hours later mice were used for imaging (n = 3–4) and biodistribution (n = 6–8) studies. The PET imaging studies were performed in 0.06, 0.6, and 3.2 mg/kg dose cohorts. To evaluate time effect on the unoccupied fraction of PD-L1 in the tumor after treatment, MDAMB231 tumor–bearing mice were treated with a single intravenous bolus dose of AtzMab (0.06, 1, and 20 mg/kg), and then imaging (n = 3) and biodistribution studies (n = 5–8) were conducted at 24 and 120 hours after AtzMab treatment. The PET imaging studies were performed in 0.06 mg/kg dose cohorts.
PET-CT imaging of mouse xenografts. Mice treated for 24 hours with therapeutic mAbs, or saline formulation as a control, were injected with 200 μCi of [64Cu]WL12 in 200 μl of formulation intravenously, and PET images were acquired 2 hours after the injection of the radiotracer. For imaging, anesthesia was induced with 3% isoflurane, and maintained with 1% isoflurane. Images were acquired in 2 bed positions at 7 min/bed in an ARGUS small-animal PET/CT scanner (Sedecal). A CT scan (512 projections) was performed at the end of each PET scan for anatomical coregistration. The PET data were reconstructed using the 2-dimensional ordered subsets-expectation maximization algorithm (2D-OSEM), and corrected for dead time and radioactive decay. The %ID per cc values were calculated based on a calibration factor obtained from a known radioactive quantity. Image fusion, visualization, and 3D rendering were accomplished using Amira 6.1 (FEI).
Ex vivo biodistribution. Mice harboring tumors and treated with therapeutic mAbs, or saline as a control (n = 5–8), were injected intravenously with 20 μCi of [64Cu]WL12. At 2 hours after [64Cu]WL12 injection, blood, tumors, and selected tissues (heart, lungs, liver, spleen, kidney, muscle) were harvested, weighed, and counted in an automated gamma counter (PerkinElmer, 2480 Wizard2 automatic gamma counter). Values for percentage of injected dose per gram of tissue (%ID/g) were calculated based on signal decay correction, and normalization to external [64Cu] standards; measurements were made in triplicate. The biodistribution data shown are presented as mean ± the standard error of the mean (SEM).
Relationship between PD-L1 mAb dose and PD-L1 engagement. To study the relationship between the AtzMab dose and unoccupied PD-L1 in the tumor, 13 dose levels were tested (0, 0.0009, 0.002, 0.006, 0.019, 0.06, 0.19, 0.6, 1.2, 3.6, 6, 12, and 24 mg/kg) in MDAMB231 tumor–bearing mice. The numbers of mice tested were the following: 9 mice for anti–PD-L1 doses of 0 mg/kg and 1.2 mg/kg (pooled from 2 repeats), 5 mice for 0.0009 mg/kg to 0.6 mg/kg, and 4 mice for 3.6 to 24 mg/kg. Each mouse first received a single dose of AtzMab and 24 hours later was injected with [64Cu]WL12. The radioactivity, measured 2 hours after the radiotracer injection, represented the unoccupied PD-L1 remaining in the tumor site, and is expressed as %ID/g.
An inhibitory sigmoidal Emax model was used to describe the effect of AtzMab dose on the unoccupied PD-L1 measured at the tumor site. The decline in unoccupied PD-L1 in the presence of AtzMab was modeled as a proportional decrease from the baseline of unoccupied PD-L1 measured in the tumor in the absence of drug, as follows: Baselineunoccupied PD-L1 is the baseline-measured unoccupied PD-L1 (in %ID/g) in the absence of AtzMab (0 mg/kg dose); Imax is the maximum inhibitory effect, i.e., the maximum fractional decrease in unoccupied PD-L1 from Baselineunoccupied PD-L1 (0 < Imax ≤ 1); ID50 is the AtzMab dose responsible for 50% of the maximum inhibitory effect (Imax/2); and Hill is the Hill coefficient describing the slope or steepness of the relationship between the AtzMab dose and the unoccupied PD-L1.
The use of a proportional inhibitory Emax model, to describe the relationship between AtzMab dose and unoccupied PD-L1 in the tumor, was considered more physiological than a conventional simple additive inhibitory Emax model, where the inhibitory effect of a drug is constant and assumed independent of the extent of expression of the target or ligand at baseline. A naive-pooled approach was used to fit the data with the proportional inhibitory sigmoidal Emax model described above. All observations were treated as being measured from a single mouse, with repeated measurement at each dose level, because of the small range and variability in body weight of the studied mice, and that each mouse contributed only one unoccupied–PD-L1 measurement.
The statistical model used to describe the residual error variability in unoccupied–PD-L1 measurements (pharmacodynamic response) was an additive error model of the following form: Observed unoccupied PD-L1 is the measured radioactivity or pharmacodynamic response; Predicted unoccupied PD-L1 is the model-predicted pharmacodynamic response; and epsilon is the residual error term, assumed to follow a normal distribution with a mean zero and variance σ2.
The developed model was used to determine ID50 of the AtzMab used in this study, and to derive the ID90 and ID96, responsible for 90% and 96% of Imax, respectively. The expected approximate average plasma concentrations corresponding to each of the ID50, ID90, and ID96 doses were also tentatively derived by dividing each dose by the steady-state volume of distribution (Vss) of the anti–PD-L1 mAb in mice. Although the actual Vss of the studied AtzMab was unknown (no pharmacokinetic study was conducted), it was assumed to be close to the reported Vss of 42.6 to 57.5 ml/kg, estimated in BALB/c mice for the chimeric anti–PD-L1 antibody PRO304397 (mouse IgG2a antibody containing the binding region of atezolizumab).
Statistics. Statistical analyses were performed using GraphPad Prism 6 software. Unpaired 2-tailed Student’s t test, 1-way ANOVA, and 2-way ANOVA were utilized for column, multiple-column, and grouped analyses, respectively. Data represent mean ± SEM. P values < 0.05 were considered statistically significant. AtzMab dose and PD-L1 engagement modeling was conducted using the nonlinear mixed-effects modeling program NONMEM (ICON Development Solutions).
Study approval. All mouse studies were conducted through Johns Hopkins University IACUC-approved protocols.
-
Author contributions
DK and AL designed the research studies, conducted radiochemistry and in vivo experiments, analyzed data, and assisted with the preparation of the manuscript. ED, MM, BW, SS, SC, WGL, PSS, and ET conducted in silico, in vitro, and ex vivo experiments, and acquired and analyzed data. VA, JTP, and CMR provided reagents. MGP, EMJ, EG, and JVSG provided technical and conceptual advice. SN conceived the idea, designed research studies, conducted experiments, analyzed data, prepared the manuscript, and supervised the study. All authors contributed to the writing and review of the manuscript.
-
Acknowledgments
We would like thank the University of Wisconsin and Washington University Cyclotron teams for 64CuCl2 production. Funding for this study was provided by Allegheny Health Network-Johns Hopkins Cancer Research Fund (to SN), NIH 1R01CA236616 (to SN), and NIH P41EB024495 (to MGP). Core resources (flow cytometry, histology, and imaging) were supported by NIH P30CA006973.
Address correspondence to: Sridhar Nimmagadda, Johns Hopkins Medical Institutions, 1550 Orleans Street, CRB II, Rm 491, Baltimore, Maryland 21287, USA. Phone: 410.502.6244; Email: snimmag1@jhmi.edu.
-
Footnotes
Conflict of interest: DK, SC, WGL, MGP, and SN are co-inventors on a pending US patent (PCT/US2017/068025) covering [64Cu]WL12 and as such are entitled to a portion of any licensing fees and royalties generated by this technology. This arrangement has been reviewed and approved by Johns Hopkins University in accordance with its conflict-of-interest policies. DK, SC, WGL, SN, JTP, CMR, and MGP are inventors on patent applications not related to this work. CMR serves on the Scientific Advisory Board of Harpoon Therapeutics, and has consulted on oncology drug development for AbbVie, Amgen, Ascentage, BMS, Celgene, Daiichi Sankyo, Genentech, Loxo, and Pharmamar. Under a licensing agreement between EMJ, Aduro Biotech Inc., and Johns Hopkins University, the University is entitled to milestone payments and royalties on sales of certain cancer vaccine products. EMJ serves on the Scientific Advisory Board of Genocea, Adaptive Biotech, DragonFly, and CSTONE and received grants from Aduro Biotech, Amgen, BMS, Hertix, and Corvus.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(2):616–630.https://doi.org/10.1172/JCI122216.
-
References
- Arrowsmith J. Trial watch: Phase II failures: 2008-2010. Nat Rev Drug Discov. 2011;10(5):328–329.View this article via: PubMed Google Scholar
- Arrowsmith J, Miller P. Trial watch: phase II and phase III attrition rates 2011-2012. Nat Rev Drug Discov. 2013;12(8):569.
- Morgan P, et al. Can the flow of medicines be improved? Fundamental pharmacokinetic and pharmacological principles toward improving Phase II survival. Drug Discov Today. 2012;17(9–10):419–424.View this article via: PubMed Google Scholar
- Jesudason CD, DuBois S, Johnson M, Barth VN, Need AB. In vivo receptor occupancy in rodents by LC-MS/MS. In: Sittampalam GS, et al, eds. Assay Guidance Manual. Bethesda,MD: Eli Lilly & Company and the National Center for Advancing Translational Sciences; 2004. View this article via: PubMed Google Scholar
- Martinez Molina D, et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341(6141):84–87.
- Dubach JM, et al. Quantitating drug-target engagement in single cells in vitro and in vivo. Nat Chem Biol. 2017;13(2):168–173.
- Burr ML, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549(7670):101–105.
- Mezzadra R, et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549(7670):106–110.
- Coelho MA, et al. Oncogenic RAS signaling promotes tumor immunoresistance by stabilizing PD-L1 mRNA. Immunity. 2017;47(6):1083–1099.e6.
- Willmann JK, van Bruggen N, Dinkelborg LM, Gambhir SS. Molecular imaging in drug development. Nat Rev Drug Discov. 2008;7(7):591–607.
- Rathkopf DE, et al. Phase I study of ARN-509, a novel antiandrogen, in the treatment of castration-resistant prostate cancer. J Clin Oncol. 2013;31(28):3525–3530.
- Nishino M, Ramaiya NH, Hatabu H, Hodi FS. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat Rev Clin Oncol. 2017;14(11):655–668.
- Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377(25):2500–2501.
- Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168(4):707–723.
- Syn NL, Teng MWL, Mok TSK, Soo RA. De-novo and acquired resistance to immune checkpoint targeting. Lancet Oncol. 2017;18(12):e731–e741.
- Hegde PS, Karanikas V, Evers S. The where, the when, and the how of immune monitoring for cancer immunotherapies in the era of checkpoint inhibition. Clin Cancer Res. 2016;22(8):1865–1874.
- Agoram BM. Use of pharmacokinetic/ pharmacodynamic modelling for starting dose selection in first-in-human trials of high-risk biologics. Br J Clin Pharmacol. 2009;67(2):153–160.
- Brahmer JR, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465.
- Lamberts LE, et al. Antibody positron emission tomography imaging in anticancer drug development. J Clin Oncol. 2015;33(13):1491–1504.
- Chatterjee S, et al. Rapid PD-L1 detection in tumors with PET using a highly specific peptide. Biochem Biophys Res Commun. 2017;483(1):258–263.
- Magiera-Mularz K, et al. Bioactive macrocyclic inhibitors of the PD-1/PD-L1 immune checkpoint. Angew Chem Int Ed Engl. 2017;56(44):13732–13735.
- Chatterjee S, et al. A humanized antibody for imaging immune checkpoint ligand PD-L1 expression in tumors. Oncotarget. 2016;7(9):10215–10227.View this article via: PubMed Google Scholar
- Mansfield AS, Dong H. Implications of programmed cell death 1 ligand 1 heterogeneity in the selection of patients with non-small cell lung cancer to receive immunotherapy. Clin Pharmacol Ther. 2016;100(3):220–222.
- Taube JM, et al. Differential expression of immune-regulatory genes associated with PD-L1 display in melanoma: Implications for PD-1 pathway blockade. Clin Cancer Res. 2015;21(17):3969–3976.
- Taube JM, et al. Colocalization of inflammatory response with B7-H1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4(127):127ra37. View this article via: PubMed Google Scholar
- Meerbrey KL, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci U S A. 2011;108(9):3665–3670.
- Heskamp S, et al. Noninvasive imaging of tumor PD-L1 expression using radiolabeled anti-PD-L1 antibodies. Cancer Res. 2015;75(14):2928–2936.
- Maute RL, et al. Engineering high-affinity PD-1 variants for optimized immunotherapy and immuno-PET imaging. Proc Natl Acad Sci U S A. 2015;112(47):E6506–E6514.
- Deng R, et al. Preclinical pharmacokinetics, pharmacodynamics, tissue distribution, and tumor penetration of anti-PD-L1 monoclonal antibody, an immune checkpoint inhibitor. MAbs. 2016;8(3):593–603.
- Hettich M, Braun F, Bartholomä MD, Schirmbeck R, Niedermann G. High-resolution PET imaging with therapeutic antibody-based PD-1/PD-L1 checkpoint tracers. Theranostics. 2016;6(10):1629–1640.
- Josefsson A, et al. Imaging, biodistribution, and dosimetry of radionuclide-labeled PD-L1 antibody in an immunocompetent mouse model of breast cancer. Cancer Res. 2016;76(2):472–479.
- Tang J, Shalabi A, Hubbard-Lucey VM. Comprehensive analysis of the clinical immuno-oncology landscape. Ann Oncol. 2018;29(1):84–91.
- Tan S, et al. Distinct PD-L1 binding characteristics of therapeutic monoclonal antibody durvalumab. Protein Cell. 2018;9(1):135–139.
- Herbst RS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567.
- Baxter LT, Jain RK. Transport of fluid and macromolecules in tumors. I. Role of interstitial pressure and convection. Microvasc Res. 1989;37(1):77–104.
- Baxter LT, Jain RK. Transport of fluid and macromolecules in tumors. II. Role of heterogeneous perfusion and lymphatics. Microvasc Res. 1990;40(2):246–263.
- Shah DK, Betts AM. Antibody biodistribution coefficients: inferring tissue concentrations of monoclonal antibodies based on the plasma concentrations in several preclinical species and human. MAbs. 2013;5(2):297–305.
- Tabrizi MA, Tseng CM, Roskos LK. Elimination mechanisms of therapeutic monoclonal antibodies. Drug Discov Today. 2006;11(1–2):81–88.View this article via: PubMed Google Scholar
- Stroh M, et al. Clinical pharmacokinetics and pharmacodynamics of atezolizumab in metastatic urothelial carcinoma. Clin Pharmacol Ther. 2017;102(2):305–312.
- Lindauer A, et al. Translational pharmacokinetic/pharmacodynamic modeling of tumor growth inhibition supports dose-range selection of the anti-PD-1 antibody pembrolizumab. CPT Pharmacometrics Syst Pharmacol. 2017;6(1):11–20.
- Lee CM, Farde L. Using positron emission tomography to facilitate CNS drug development. Trends Pharmacol Sci. 2006;27(6):310–316.
- Sheng J, et al. Clinical Pharmacology considerations for the development of immune checkpoint inhibitors. J Clin Pharmacol. 2017;57(Suppl 10):S26–S42.View this article via: PubMed Google Scholar
- Biotech PhD. The strange tale of pidilizumab. The Science of Biotech. https://www.scienceofbiotech.com/strange-tale-pidilizumab/ Published March 10, 2017. Accessed November 27, 2018.
- Roach C, et al. Development of a companion diagnostic PD-L1 immunohistochemistry assay for pembrolizumab therapy in non-small-cell lung cancer. Appl Immunohistochem Mol Morphol. 2016;24(6):392–397.
- Meng X, Huang Z, Teng F, Xing L, Yu J. Predictive biomarkers in PD-1/PD-L1 checkpoint blockade immunotherapy. Cancer Treat Rev. 2015;41(10):868–876.
- Ilie M, et al. Comparative study of the PD-L1 status between surgically resected specimens and matched biopsies of NSCLC patients reveal major discordances: a potential issue for anti-PD-L1 therapeutic strategies. Ann Oncol. 2016;27(1):147–153.
- McLaughlin J, et al. Quantitative assessment of the heterogeneity of PD-L1 expression in non-small-cell lung cancer. JAMA Oncol. 2016;2(1):46–54.
- Nolan E, et al. Combined immune checkpoint blockade as a therapeutic strategy for BRCA1-mutated breast cancer. Sci Transl Med. 2017;9(393):eaal4922.
- Gniadek TJ, Li QK, Tully E, Chatterjee S, Nimmagadda S, Gabrielson E. Heterogeneous expression of PD-L1 in pulmonary squamous cell carcinoma and adenocarcinoma: implications for assessment by small biopsy. Mod Pathol. 2017;30(4):530–538.
- Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016;17(12):e542–e551.
- Weissleder R, Schwaiger MC, Gambhir SS, Hricak H. Imaging approaches to optimize molecular therapies. Sci Transl Med. 2016;8(355):355ps16.
- Ascierto ML, et al. Transcriptional mechanisms of resistance to anti-PD-1 therapy. Clin Cancer Res. 2017;23(12):3168–3180.
- Netti PA, Berk DA, Swartz MA, Grodzinsky AJ, Jain RK. Role of extracellular matrix assembly in interstitial transport in solid tumors. Cancer Res. 2000;60(9):2497–2503.View this article via: PubMed Google Scholar
- Kamath AV. Translational pharmacokinetics and pharmacodynamics of monoclonal antibodies. Drug Discov Today Technol. 2016;21–22:75–83.View this article via: PubMed Google Scholar
- Azad BB, et al. A fully human CXCR4 antibody demonstrates diagnostic utility and therapeutic efficacy in solid tumor xenografts. Oncotarget. 2016;7(11):12344–12358.View this article via: PubMed Google Scholar
- Powles T, et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515(7528):558–562.
- Zak KM, et al. Structure of the complex of human programmed death 1, PD-1, and its ligand PD-L1. Structure. 2015;23(12):2341–2348.
- Liu K, et al. Structural basis of anti-PD-L1 monoclonal antibody avelumab for tumor therapy. Cell Res. 2017;27(1):151–153.
- Lee HT, et al. Molecular mechanism of PD-1/PD-L1 blockade via anti-PD-L1 antibodies atezolizumab and durvalumab. Sci Rep. 2017;7(1):5532.
- Lee JY, et al. Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy. Nat Commun. 2016;7:13354. View this article via: PubMed Google Scholar
- Zhang F, et al. Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade. Cell Discov. 2017;3:17004. View this article via: PubMed Google Scholar
- Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242.
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38, 27–28.View this article via: PubMed Google Scholar
- Yang X, et al. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8(8):659–661.
- Arrowsmith J. Trial watch: Phase II failures: 2008-2010. Nat Rev Drug Discov. 2011;10(5):328–329.
-
Version history
- Version 1 (November 20, 2018): In-Press Preview
- Version 2 (January 7, 2019): Electronic publication
- Version 3 (February 1, 2019): Print issue publication



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)

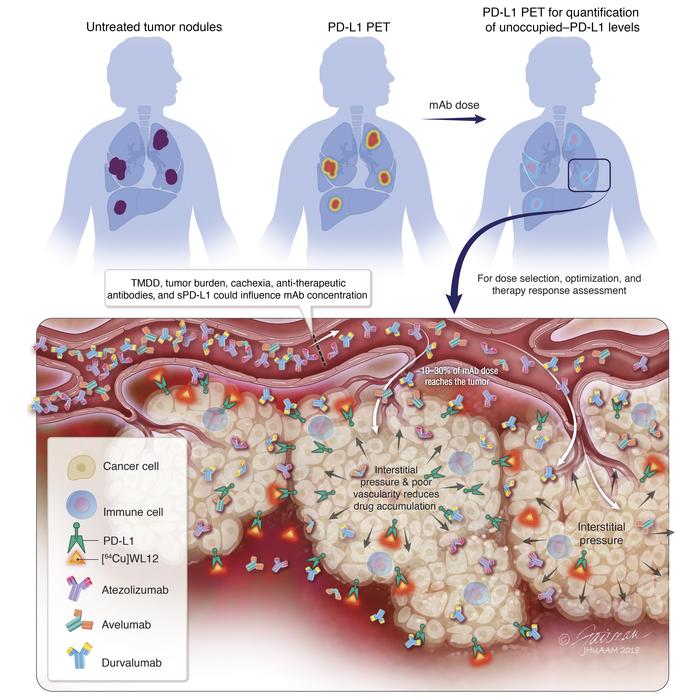



![PD-L1 engagement by anti–PD-L1 mAbs is quantified at the tumor using [64Cu]WL12 in xenografts with variable PD-L1 expression. PD-L1 engagement by anti–PD-L1 mAbs is quantified at the tumor using [64Cu]](http://dm5migu4zj3pb.cloudfront.net/manuscripts/122000/122216/small/JCI122216.f3.gif)
![Dynamic changes in tumor PD-L1 expression and its engagement by AtzMab detected using [64Cu]WL12. Dynamic changes in tumor PD-L1 expression and its engagement by AtzMab dete](http://dm5migu4zj3pb.cloudfront.net/manuscripts/122000/122216/small/JCI122216.f4.gif)
![Tumor PD-L1 engagement by 3 distinct PD-L1 therapeutic antibodies quantified using [64Cu]WL12. Tumor PD-L1 engagement by 3 distinct PD-L1 therapeutic antibodies quantifie](http://dm5migu4zj3pb.cloudfront.net/manuscripts/122000/122216/small/JCI122216.f5.gif)
![Effect of dose and time on the unoccupied fraction of tumor PD-L1 following treatment with AtzMab quantified using [64Cu]WL12. Effect of dose and time on the unoccupied fraction of tumor PD-L1 following](http://dm5migu4zj3pb.cloudfront.net/manuscripts/122000/122216/small/JCI122216.f6.gif)
![Effect of dose and time on tumor PD-L1 occupancy by AtzMab quantified using [64Cu]WL12. Effect of dose and time on tumor PD-L1 occupancy by AtzMab quantified using](http://dm5migu4zj3pb.cloudfront.net/manuscripts/122000/122216/small/JCI122216.f7.gif)
