Advertisement
Concise CommunicationOncology Free access | 10.1172/JCI92710
Hedgehog signaling drives medulloblastoma growth via CDK6
David R. Raleigh,1,2 Pervinder K. Choksi,1,2 Alexis Leigh Krup,1,2 Wasima Mayer,1,2 Nicole Santos,2 and Jeremy F. Reiter2
1Department of Radiation Oncology and
2Department of Biochemistry and Biophysics, UCSF, San Francisco, California, USA.
Address correspondence to: Jeremy F. Reiter, Department of Biochemistry, University of California San Francisco, 555 Mission Bay Boulevard South, CVRI Building, Room 384S, San Francisco, California 94158, USA. Phone: 415.476.0156; Email: jeremy.reiter@ucsf.edu.
Find articles by Raleigh, D. in: JCI | PubMed | Google Scholar
1Department of Radiation Oncology and
2Department of Biochemistry and Biophysics, UCSF, San Francisco, California, USA.
Address correspondence to: Jeremy F. Reiter, Department of Biochemistry, University of California San Francisco, 555 Mission Bay Boulevard South, CVRI Building, Room 384S, San Francisco, California 94158, USA. Phone: 415.476.0156; Email: jeremy.reiter@ucsf.edu.
Find articles by Choksi, P. in: JCI | PubMed | Google Scholar
1Department of Radiation Oncology and
2Department of Biochemistry and Biophysics, UCSF, San Francisco, California, USA.
Address correspondence to: Jeremy F. Reiter, Department of Biochemistry, University of California San Francisco, 555 Mission Bay Boulevard South, CVRI Building, Room 384S, San Francisco, California 94158, USA. Phone: 415.476.0156; Email: jeremy.reiter@ucsf.edu.
Find articles by Krup, A. in: JCI | PubMed | Google Scholar
1Department of Radiation Oncology and
2Department of Biochemistry and Biophysics, UCSF, San Francisco, California, USA.
Address correspondence to: Jeremy F. Reiter, Department of Biochemistry, University of California San Francisco, 555 Mission Bay Boulevard South, CVRI Building, Room 384S, San Francisco, California 94158, USA. Phone: 415.476.0156; Email: jeremy.reiter@ucsf.edu.
Find articles by Mayer, W. in: JCI | PubMed | Google Scholar
1Department of Radiation Oncology and
2Department of Biochemistry and Biophysics, UCSF, San Francisco, California, USA.
Address correspondence to: Jeremy F. Reiter, Department of Biochemistry, University of California San Francisco, 555 Mission Bay Boulevard South, CVRI Building, Room 384S, San Francisco, California 94158, USA. Phone: 415.476.0156; Email: jeremy.reiter@ucsf.edu.
Find articles by Santos, N. in: JCI | PubMed | Google Scholar
1Department of Radiation Oncology and
2Department of Biochemistry and Biophysics, UCSF, San Francisco, California, USA.
Address correspondence to: Jeremy F. Reiter, Department of Biochemistry, University of California San Francisco, 555 Mission Bay Boulevard South, CVRI Building, Room 384S, San Francisco, California 94158, USA. Phone: 415.476.0156; Email: jeremy.reiter@ucsf.edu.
Find articles by Reiter, J. in: JCI | PubMed | Google Scholar
Published November 20, 2017 - More info
J Clin Invest. 2018;128(1):120–124. https://doi.org/10.1172/JCI92710.
Copyright © 2018, American Society for Clinical Investigation
Received: January 9, 2017; Accepted: October 12, 2017
-
Abstract
Medulloblastoma, an aggressive cancer of the cerebellum, is among the most common pediatric brain tumors. Approximately one-third of medulloblastomas are associated with misactivation of the Hedgehog (Hh) pathway. GLI family zinc finger 2 (GLI2) coordinates the Hh transcriptional program; however, the GLI2 targets that promote cancer cell proliferation are unknown. Here, we incorporated a Gli2-EGFP allele into 2 different genetic mouse models of Hh-associated medulloblastoma. Hh signaling induced GLI2 binding to the Cdk6 promoter and activated Cdk6 expression, thereby promoting uncontrolled cell proliferation. Genetic or pharmacological inhibition of CDK6 in mice repressed the growth of Hh-associated medulloblastoma and prolonged survival through inhibition of cell proliferation. In human medulloblastoma, misactivation of Hh signaling was associated with high levels of CDK6, pointing to CDK6 as a direct transcriptional target of the Hh pathway. These results suggest that CDK6 antagonists may be a promising therapeutic approach for Hh-associated medulloblastoma in humans.
-
Introduction
Medulloblastoma, an aggressive cancer of the cerebellum, is among the most common pediatric brain tumors (1). Transcriptional profiling studies reveal that medulloblastomas exist as 4 main molecular subgroups (2). Approximately one-third of medulloblastomas are associated with misactivation of the Hedgehog (Hh) pathway, a signal transduction pathway that is essential for development (3). Vertebrate Hh signals are transduced through the primary cilium, an antenna that projects from the surface of most cells. Cells of the cerebellar external granule layer (EGL) give rise to Hh-associated medulloblastoma and are ciliated (4, 5). Other Hh-related cancer cells, such as basal cell carcinoma cells, are also ciliated, and disrupting either cilia or ciliary Hh signaling blocks cancer growth in both basal cell carcinoma and medulloblastoma (5, 6).
Hh ligands relieve Patched1 (PTCH1) repression of Smoothened (SMO), allowing SMO to localize to cilia and activate GLI family zinc finger 2 (GLI2), the principle effector of the Hh transcriptional program (3). The targets of GLI2 that drive uncontrolled cell proliferation in cancer are poorly understood. Here, we demonstrate that GLI2 binds to the Cdk6 promoter to induce cell proliferation in response to Hh signals. Inhibiting CDK6 blocks the growth of Hh-associated medulloblastoma in vivo, suggesting that pharmacologic inhibition of CDK6 may be an effective strategy for patients with Hh-associated cancers.
-
Results and Discussion
To study how misactivation of GLI2 causes cancer, we used the Floxin system to generate a Gli2-knockin allele that encodes a fusion of GLI2 to EGFP and FLAG tags (Gli2-EGFP) (7). Mice homozygous for the Gli2-EGFP allele are viable and morphologically indistinguishable from WT, revealing that this fusion protein is functional (8). To investigate the function of GLI2 in medulloblastoma, we incorporated the Gli2-EGFP allele into 2 established mouse models of Hh-associated medulloblastoma. These models make use of Cre recombinase under the control of Math1 regulatory sequences to express a constitutively active, oncogenic point mutant of Smo (SmoM2c) or to inactivate Ptch1 (Ptch1c) in the EGL (9, 10). In these mouse models, GLI2-EGFP (i) was expressed under endogenous regulatory control, (ii) recapitulated GLI2 activity, interactions, and localization, and (iii) allowed us to immunoprecipitate GLI2 and identify target genes (8).
RNA sequencing of Math1-Cre SmoM2 Gli2-EGFP medulloblastomas demonstrated that, as expected, general Hh target genes, such as Ptch1, Ptch2, and Gli1, and recognized markers of Hh-associated medulloblastoma, including Sfrp1, Pdlim3, and Met, were increased relative to control cerebellums (Figure 1A, Supplemental Figure 1, A and B, and Supplemental Table 1). Other genes not involved in Hh signaling itself were also upregulated, including Cdk6, the expression of which was 167 ± 42–fold higher than that in controls (Figure 1, A and B). Cdk6 encodes cell division kinase 6 (CDK6), which, when bound to cyclin D, phosphorylates retinoblastoma protein (RB) and activates E2F transcription factors to stimulate cell-cycle progression. Like Cdk6 transcript, CDK6 protein was strongly increased in Hh-associated medulloblastoma (Figure 1C). As (i) Cdk6 is a target of Hh signaling during limb-bud development, (ii) expression of CDK6 is an independent negative prognostic factor in human medulloblastoma, and (iii) inhibition of CDK6 in vitro suppresses medulloblastoma cell proliferation, we hypothesized that CDK6 could be functionally important in Hh pathway–associated medulloblastoma (11–13). Therefore, we compared CDK6 expression in diverse adult and pediatric human brain tumors and found that CDK6 was particularly elevated in medulloblastoma (Supplemental Figure 1, C and D). Interestingly, CDK6 expression was equivalent in all 4 classes of medulloblastoma, suggesting that it may be a common effector of uncontrolled cell proliferation in medulloblastoma regardless of genetic etiology (Supplemental Figure 1E). In support of this hypothesis, small molecule inhibition of CDK6 confers a survival benefit in mice bearing patient-derived xenographs of group 3 medulloblastomas (14).
 Figure 1
Figure 1Hh signaling induces CDK6 in medulloblastoma. (A) Volcano plot generated by RNA sequencing of the medulloblastomas from 3 P35 Math1-Cre SmoM2c Gli2-EGFP female mice relative to the cerebella of 3 P35 SmoM2c Gli2-EGFP female littermate controls. 2676 genes are differentially expressed with fold-change of 3 or more, P value of less than 0.008, and false discovery rate of 0.05 or less. Hh pathway targets and Cdk6 (red) are enriched in Hh-associated medulloblastoma. (B) Quantitative reverse-transcriptase PCR (qRT-PCR) demonstrates enrichment of Gli1 and enrichment of Cdk6 expression in Hh-associated medulloblastoma (blue) relative to age- and sex-matched littermate cerebella (gray). *P < 0.004, t test. n = 4 mice per genotype, representative of 3 experiments. (C) Immunoblot demonstrates elevated levels of CDK6, cyclin D1, and phospho-RB proteins in Hh-associated medulloblastoma. n = 2 mice, representative of 3 experiments. (D) qRT-PCR of ciliated NIH/3T3 cells treated either with SAG alone to activate the Hh pathway (green) or in conjunction with HLM006474 to inhibit E2F DNA binding (black). E2F antagonism inhibits induction of all cell cycle effectors in response to Hh stimulation except for Cdk6 and Ccnd1. *P < 0.05, t test. n = 3, representative of 3 experiments.
The levels of the CDK6-interacting cyclin cyclin D1 were also elevated in Hh pathway–associated medulloblastoma (Figure 1C, Supplemental Table 1, and Supplemental Figure 2A). We therefore assessed phosphorylated RB levels and found them to be dramatically increased in Hh pathway–associated medulloblastoma (Figure 1C). Consistently, medulloblastomas displayed markedly elevated expression of E2F target genes (Supplemental Figure 2A), further suggesting that misactivation of Hh signaling may drive cell cycle progression via CDK6 (15). Of note, the read count of the related mitogenic kinase Cdk4 was higher than that of Cdk6 in Hh-associated medulloblastoma, but the differential expression of Cdk4 in Hh-associated medulloblastoma relative to normal cerebellum was 13.9-fold less than that of Cdk6 (Supplemental Figure 2, A and B, and Supplemental Table 1).
In human medulloblastoma, CDK6 expression was elevated even relative to that in highly proliferative neural progenitors (Supplemental Figure 2C), further raising the possibility that misactivation of Hh signaling induces super-physiological levels of Cdk6 expression. To test whether Hh signals are sufficient to induce Cdk6 expression, we activated NIH/3T3 cells, a Hh-responsive cell type, with smoothened agonist (SAG) (16). Hh pathway activation in this system induced a modest increase in expression of cell cycle effectors, including Ccnd1 and Cdk6 (Figure 1D and Supplemental Figure 2D). Concurrent administration of SAG and an E2F-DNA binding antagonist blocked the Hh pathway–mediated induction of all cell cycle transcripts examined except for Ccnd1 and Cdk6 (Figure 1D). Concurrent inhibition of translation blocked the induction of Ccnd1 in response to Hh stimulation, but did not affect Cdk6 induction (Figure 2A). These data suggest that, whereas Hh signaling affects the expression of diverse cell cycle effectors, Cdk6 is exceptional in that it is a direct transcriptional target of the pathway.
 Figure 2
Figure 2GLI2 binds the Cdk6 promoter to activate gene expression. (A) qRT-PCR of NIH/3T3 cells treated vehicle (gray), SAG alone (green) or in combination with the translation inhibitor cycloheximide (black). *P < 0.02, t test. n = 3, representative of 3 experiments. (B) Schematic of Cdk6 promoter showing putative GLI-binding sites. (C) EGFP ChIP-qPCR from 3 Math1-Cre SmoM2c Gli2-EGFP tumors (blue) and 3 age- and sex-matched SmoM2c Gli2-EGFP littermate cerebella (gray). *P < 0.03, t test. Representative of 3 experiments. (D) Histone ChIP-qPCR from 3 Math1-Cre SmoM2c tumors (aqua) and 3 age- and sex-matched SmoM2c littermate control cerebella (gray). *P < 0.002, t test. Representative of 2 experiments. (E) EGFP ChIP-qPCR from NIH/3T3 cells stably expressing GLI2-EGFP treated with vehicle (gray), SAG alone (green), or SAG in combination with the SMO inhibitor vismodegib (black). *P < 0.008, t test. n = 3, representative of 3 experiments. (F) Activity of the Cdk6 promoter luciferase reporter in NIH/3T3 cells treated with vehicle (gray) or SAG (green). SAG activates the Cdk6 promoter extending from GLI2-binding site 1 to the transcriptional start site (1-Start) or from site 4 to the transcriptional start site (4-Start), but not from site 1 to site 3 (s1–3). *P < 0.01, t test. n = 3, representative of 3 experiments. (G) Activity of the Cdk6 promoter luciferase reporter in NIH/3T3 cells cotransfected with control vector (gray) or a constitutively active GLI2-CLEG plasmid (green). *P < 0.02, t test. n = 3, representative of 2 experiments. (H) Activity of the Cdk6 promotor luciferase reporter in NIH/3T3 cells cotransfected with control vector (gray) or GLI2-CLEG (green) shows GLI2-CLEG activates gene expression from 3 copies of the Cdk6 promoter site 4, but not from 3 tandem mutant copies. *P < 0.001, t test. n = 3, representative of 3 experiments.
The Cdk6 promoter binds GLI3, the principle repressor of the Hh transcriptional program, during limb-bud development (11, 17). ChIP analysis of GLI2-EGFP–binding sites in medulloblastomas from Math1-Cre SmoM2c Gli2-EGFP mice revealed that, compared with cerebella from control GLI2-EGFP–expressing mice, GLI2-EGFP is selectively enhanced at a previously identified cis-regulatory element at the Cdk6 promoter that is involved in Hh signaling–mediated limb development (site 4, Figure 2, B and C, and Supplemental Figure 3, A–F) (11). Consistent with Hh pathway–associated binding of a transcriptional activator, site 4 also exhibited an enrichment of H3K27 acetylation, which is associated with transcriptional activation, and a suppression of H3K27me3, which is associated with transcriptional repression, in medulloblastoma (Figure 2D and Supplemental Figure 3G). Similarly, ChIP of NIH/3T3 cells expressing EGFP-GLI2 showed increased occupancy of EGFP-GLI2 at site 4 upon activation of Hh signaling (Figure 2E and Supplemental Figure 3, H–J).
To test whether GLI2 binding to the Cdk6 promoter is sufficient to activate transcription, we made luciferase reporter constructs containing truncated Cdk6 promoter sequences. Transfection of reporters into NIH/3T3 cells revealed that Cdk6 promoter sequences that included site 4 were sufficient to confer responsiveness to SAG-mediated activation of the Hh pathway (Figure 2F). Similarly, Cdk6 promoter sequences that included site 4 conferred responsiveness to a constitutively active form of GLI2, GLI2-CLEG (Figure 2G). In contrast, a Cdk6 promoter sequence that did not include site 4 was not responsive to either SAG or GLI2-CLEG (Figure 2, F and G). Furthermore, multimerized site 4 without surrounding Cdk6 sequences was sufficient to confer transcriptional responsiveness to GLI2-CLEG (Figure 2H and Supplemental Table 2).
Cdk6-null mice have no overt developmental phenotypes, and Cdk6 is not required for cerebellar development (Supplemental Figure 4, A and B) (18). To test whether CDK6 is functionally important for Hh-associated cancer growth, we genetically removed CDK6 from the Math1-Cre SmoM2c medulloblastoma model. Homozygous genetic deletion of Cdk6 (Math1-Cre SmoM2c Cdk6KO/KO) reduced the weight (32% ± 2%) and size of tumors relative to those with either 1 or 2 copies of the Cdk6 allele (Figure 3A). Homozygous genetic deletion of Cdk6 also reduced the prevalence of small round blue cells that are characteristic of medulloblastoma and partially restored cerebellar architecture (Figure 3B and Supplemental Figure 4, C and D). Moreover, genetically removing CDK6 function prolonged median survival in Math1-Cre SmoM2c animals (97 days versus 52 days) (Figure 3C). To confirm the involvement of CDK6 in Hh-associated medulloblastoma, we genetically removed CDK6 in a second tumor model, one that relies on the loss of the negative regulator of the pathway, PTCH1, rather than activation of SMO. As with the SMO misactivation tumors, homozygous genetic deletion of Cdk6 in the Math1-Cre Ptch1c/c tumors prolonged median survival relative to animals with 2 copies of the Cdk6 allele (135 versus 68 days, Figure 3D).
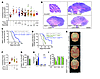 Figure 3
Figure 3Inhibition of CDK6 attenuates Hh-associated medulloblastoma growth. (A) Cerebella weight normalized to total brain weight from P35 mice (SmoM2c [n = 12, white], Math1-Cre SmoM2c [n = 12, gray], Math1-Cre SmoM2c Cdk6KO/+ [n = 7, light blue], and Math1-Cre SmoM2c Cdk6KO/KO [n = 19, dark blue]) and P35 mice treated with pharmacologic agents (Math1-Cre SmoM2c with vismodegib 75 μg/g [n = 15, black], vismodegib 100 μg/g [n = 14, black], palbociclib 50 μg/g [n = 11, green], palbociclib 100 μg/g [n = 12, green], or vismodegib 75 μg/g, and palbociclib 50 μg/g [n = 13, orange], and Math1-Cre SmoM2c Cdk6KO/KO with palbociclib 100 μg/g [n = 6, blue]). P < 0.04, t test. (B) P35 sagittal midline cerebellar H&E light micrographs. Scale bar: 1 mm (representative of 3 experiments). (C) Kaplan-Meier curves of 20 Math1-Cre SmoM2c (gray), 24 Math1-Cre SmoM2c Cdk6KO/+ (light blue), and 25 Math1-Cre SmoM2c Cdk6KO/KO mice. P < 0.0001, log-rank test. (D) Kaplan-Meier curves of 13 Math1-Cre Ptch1c/c Cdk6+/+ (gray), 10 Math1-Cre Ptch1c/c Cdk6KO/KO (blue), and 8 Math1-Cre Ptch1c/c Cdk6+/+ mice treated with 100 μg/g palbociclib from P21 until death (green). P < 0.0001, log-rank test. (E) P35 brain micrographs. Scale bar: 5 mm (representative of 3 experiments). (F) Cerebella weight normalized to total brain weight of P35 mice, including 12 Ptch1c/c (white), 12 Math1-Cre Ptch1c/c (gray), and Math1-Cre Ptch1c/c, treated with 75 μg/g abemaciclib (n = 8, light green) or 100 μg/g palbociclib (n = 11, green). (G) BrdU quantification of Math1-Cre SmoM2c medulloblastomas. P < 0.03, t test. n = 3. (H) qRT-PCR of DAOY, D283, and D341 cells. P < 0.05, t test. n = 3. (I) Ki-67 quantification in DAOY, D283, and D341 cells. *P < 0.05, t test. n = 6.
We hypothesized that, like genetic deletion of Cdk6, pharmacological inhibition of CDK6 would inhibit the growth of medulloblastoma. To test this hypothesis, we treated Math1-Cre SmoM2c and Math1-Cre Ptch1c/c mice with a small molecule inhibitor of CDK4/6, palbociclib. A positive control, the SMO inhibitor vismodegib, reduced medulloblastoma weight (32% ± 2%, Figure 3A) (19). Pharmacologic inhibition of CDK4/6 with palbociclib reduced tumor weight in both Math1-Cre SmoM2c (31% ± 3%, Figure 3, A and E) and Math1-Cre Ptch1c/c (41% ± 5%, Figure 3F) mice to an extent similar to that seen with vismodegib. Much like genetic deletion of Cdk6, palbociclib also reduced the size of tumors, decreased the prevalence of small round blue cells, partially restored cerebellar architecture, and prolonged survival (Figure 3D and Supplemental Figure 4, C and D).
To confirm the efficacy of CDK6 inhibition for medulloblastoma, we treated Math1-Cre Ptch1c/c mice with a different small molecule CDK4/6 antagonist abemaciclib, which also reduced the size of tumors (14% ± 2%, Figure 3F). As both palbociclib and abemaciclib inhibit CDK4 in addition to CDK6, we treated Math1-Cre SmoM2c Cdk6KO/KO mice with palbociclib to test whether inhibition of CDK4 contributes to their effect on medulloblastoma size. We did not detect a difference in Math1-Cre SmoM2c Cdk6KO/KO tumor weight with and without palbociclib, suggesting that CDK4 is not a significant driver of Hh-associated medulloblastoma growth (Figure 3A).
Medulloblastoma acquires resistance to single-agent molecular therapy with vismodegib (19, 20). Further suggesting a possible role for CDK4/6 inhibition in medulloblastoma treatment, therapeutic doses of vismodegib are associated with premature growth plate fusion, but palbociclib is not (Supplemental Figure 4E) (21). To determine whether combination molecular therapy is an effective strategy for medulloblastoma, we treated Math1-Cre SmoM2c mice with vismodegib and palbociclib. We identified substantial morbidity with full-dose combination molecular therapy (150 μg/g vismodegib and 100 μg/g palbociclib). Thus, we reduced the dose of each agent (75 μg/g vismodegib and 50 μg/g palbociclib). Low-dose monotherapy with either agent failed to reduce tumor weights as much as full-dose treatment (vismodegib, 24% ± 2%; palbociclib, 28% ± 3%; Figure 3A). However, low-dose combination therapy reduced tumor weight more than either agent and to an extent comparable to that of full-dose monotherapy (39% ± 1%). These data suggest that simultaneous molecular inhibition of SMO and CDK6 may be an effective strategy for inhibiting the growth of Hh-associated medulloblastoma.
To understand the mechanism by which CDK4/6 inhibition attenuates the growth of Hh-associated medulloblastoma, we quantified tumor cell apoptosis and proliferation after palbociclib treatment in Math1-Cre SmoM2c mice. Pharmacologic inhibition of CDK6 had no effect on tumor apoptosis (Supplemental Figure 4, F and G). In contrast, palbociclib reduced the amount of BrdU-positive cells by 35% ± 2%, indicating that CDK4/6 inhibition diminished cell proliferation (Figure 3G and Supplemental Figure 5A). As inhibiting CDK6 induces G1 arrest and cellular senescence (22, 23), we hypothesized that the effect of CDK4/6 inhibition on Hh-associated medulloblastoma is mostly cytostatic. In support of this hypothesis, tumor cell proliferation recovered following palbociclib withdrawal (Supplemental Figure 5, B and C).
To test the generalizability of CDK4/6 inhibition for other medulloblastoma molecular subgroups, we treated diverse human medulloblastoma cell lines with palbociclib and quantified cell proliferation. DAOY medulloblastoma cells, representative of Hh-associated medulloblastoma, had elevated expression of Gli1, Ptch1, and Cdk6 relative to D283 and D341 medulloblastoma cells, which is representative of group 3 or group 4 medulloblastoma (Figure 3H) (24, 25). Consistently, palbociclib significantly reduced the amount of Ki-67–positive DAOY cells in a dose-dependent manner and only mildly reduced Ki-67 expression in D283 and D341 cells (Figure 3I and Supplemental Figure 5D). These data suggest that CDK4/6 inhibition may be most effective in medulloblastoma tumors with elevated CDK6 expression.
In conclusion, we demonstrate that misactivation of Hh signaling in cancer induces CDK6 to drive medulloblastoma growth. The main transcriptional effector of Hh signaling, GLI2, binds to a site within the Cdk6 promoter to induce CDK6. In turn, CDK6 phosphorylates RB to activate E2F and induce medulloblastoma cell proliferation. Either genetic or pharmacologic inhibition of CDK6 in 2 genetically distinct mouse models reduces medulloblastoma proliferation, reduces tumor burden, and prolongs survival. We propose that, as a direct transcriptional target of GLI2, CDK6 is a principal means by which the Hh pathway activates the cell cycle in cancer. Therefore, we hypothesize that CDK4/6 inhibition will be an effective therapy for patients with Hh-associated medulloblastoma.
-
Methods
Please see Supplemental Methods for a detailed explanation of all experimental procedures.
Study approval. Animal experiments were conducted in a Laboratory Animal Resource Center per UCSF Institutional Animal Care and Use Committee–approved protocol AN098101.
-
Author contributions
DRR designed research studies, conducted experiments, acquired data, analyzed data, and wrote the manuscript. PKC conducted experiments, acquired data, and analyzed data. ALK conducted experiments, acquired data, and analyzed data. WM conducted experiments. NS provided reagents. JFR designed research studies and wrote the manuscript.
-
Acknowledgments
This work was supported by grants from the NIH (AR054396 and GM095941) and from the Burroughs Wellcome Fund, the Packard Foundation, and the V Foundation (to JFR), and grants from the NIH (HL007731 and CA212279-01), the UCSF Physician Scientist Scholar Program, the American Society of Clinical Oncology, the Rally Foundation for Childhood Cancer Research, and the American Brain Tumor Association (to DRR).
Address correspondence to: Jeremy F. Reiter, Department of Biochemistry, University of California San Francisco, 555 Mission Bay Boulevard South, CVRI Building, Room 384S, San Francisco, California 94158, USA. Phone: 415.476.0156; Email: jeremy.reiter@ucsf.edu.
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(1):120–124.https://doi.org/10.1172/JCI92710.
-
References
- Ostrom QT, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the united states in 2008-2012. Neuro-oncology. 2015;17 Suppl 4:iv1–iv62.View this article via: PubMed Google Scholar
- Northcott PA, et al. Medulloblastomics: the end of the beginning. Nat Rev Cancer. 2012;12(12):818–834.
- Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14(7):416–429.View this article via: PubMed Google Scholar
- Schüller U, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14(2):123–134.
- Han YG, Kim HJ, Dlugosz AA, Ellison DW, Gilbertson RJ, Alvarez-Buylla A. Dual and opposing roles of primary cilia in medulloblastoma development. Nat Med. 2009;15(9):1062–1065.
- Wong SY, et al. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat Med. 2009;15(9):1055–1061.
- Singla V, et al. Floxin, a resource for genetically engineering mouse ESCs. Nat Methods. 2010;7(1):50–52.
- Santos N, Reiter JF. A central region of Gli2 regulates its localization to the primary cilium and transcriptional activity. J Cell Sci. 2014;127(Pt 7):1500–1510.View this article via: PubMed Google Scholar
- Xie J, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391(6662):90–92.
- Raffel C, et al. Sporadic medulloblastomas contain PTCH mutations. Cancer Res. 1997;57(5):842–845.View this article via: PubMed Google Scholar
- Lopez-Rios J, et al. GLI3 constrains digit number by controlling both progenitor proliferation and BMP-dependent exit to chondrogenesis. Dev Cell. 2012;22(4):837–848.
- Mendrzyk F, et al. Genomic and protein expression profiling identifies CDK6 as novel independent prognostic marker in medulloblastoma. J Clin Oncol. 2005;23(34):8853–8862.
- Whiteway SL, et al. Inhibition of cyclin-dependent kinase 6 suppresses cell proliferation and enhances radiation sensitivity in medulloblastoma cells. J Neurooncol. 2013;111(2):113–121.
- Cook Sangar ML, et al. Inhibition of CDK4/6 by palbociclib significantly extends survival in medulloblastoma patient-derived xenograft mouse models. Clin Cancer Res. 2017;23(19):5802–5813.
- Bhatia B, Malik A, Fernandez-LA , Kenney AM. p27(Kip1), a double-edged sword in Shh-mediated medulloblastoma: Tumor accelerator and suppressor. Cell Cycle. 2010;9(21):4307–4314.
- Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci U S A. 2002;99(22):14071–14076.
- Vokes SA, Ji H, Wong WH, McMahon AP. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 2008;22(19):2651–2663.
- Malumbres M, et al. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell. 2004;118(4):493–504.
- Yauch RL, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326(5952):572–574.
- Rudin CM, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361(12):1173–1178.
- Lucas JT, Wright KD. Vismodegib and physeal closure in a pediatric patient. Pediatr Blood Cancer. 2016;63(11):2058.
- Fry DW, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther. 2004;3(11):1427–1438.View this article via: PubMed Google Scholar
- Thangavel C, et al. Therapeutically activating RB: reestablishing cell cycle control in endocrine therapy-resistant breast cancer. Endocr Relat Cancer. 2011;18(3):333–345.
- Friedman HS, et al. A model for human medulloblastoma. Growth, morphology, and chromosomal analysis in vitro and in athymic mice. J Neuropathol Exp Neurol. 1983;42(5):485–503.
- Jacobsen PF, Jenkyn DJ, Papadimitriou JM. Establishment of a human medulloblastoma cell line and its heterotransplantation into nude mice. J Neuropathol Exp Neurol. 1985;44(5):472–485.
- Ostrom QT, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the united states in 2008-2012. Neuro-oncology. 2015;17 Suppl 4:iv1–iv62.
-
Version history
- Version 1 (November 20, 2017): Electronic publication
- Version 2 (December 11, 2017): Added supplemental methods.
- Version 3 (January 2, 2018): Print issue publication



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)



