Advertisement
Research ArticleOncology Free access | 10.1172/JCI43354
Targeting CDK1 promotes FLT3-activated acute myeloid leukemia differentiation through C/EBPα
Hanna S. Radomska,1 Meritxell Alberich-Jordà,1,2 Britta Will,1 David Gonzalez,1 Ruud Delwel,3 and Daniel G. Tenen2,4
1Beth Israel Deaconess Medical Center/Harvard Medical School, Boston, Massachusetts, USA. 2Harvard Stem Cell Institute, Harvard Medical School, Boston, Massachusetts, USA. 3Erasmus University, Rotterdam, Netherlands. 4Cancer Science Institute, National University of Singapore, Singapore.
Address correspondence to: Daniel G. Tenen, Center for Life Sciences, 3 Blackfan Circle, Room 437, Boston, Massachusetts 02115, USA. Phone: 617.735.2205; Fax: 617.735.2222; E-mail: dtenen@bidmc.harvard.edu.
Authorship note: Hanna S. Radomska and Meritxell Alberich-Jordà contributed equally to this work.
Find articles by Radomska, H. in: JCI | PubMed | Google Scholar
1Beth Israel Deaconess Medical Center/Harvard Medical School, Boston, Massachusetts, USA. 2Harvard Stem Cell Institute, Harvard Medical School, Boston, Massachusetts, USA. 3Erasmus University, Rotterdam, Netherlands. 4Cancer Science Institute, National University of Singapore, Singapore.
Address correspondence to: Daniel G. Tenen, Center for Life Sciences, 3 Blackfan Circle, Room 437, Boston, Massachusetts 02115, USA. Phone: 617.735.2205; Fax: 617.735.2222; E-mail: dtenen@bidmc.harvard.edu.
Authorship note: Hanna S. Radomska and Meritxell Alberich-Jordà contributed equally to this work.
Find articles by Alberich-Jordà, M. in: JCI | PubMed | Google Scholar
1Beth Israel Deaconess Medical Center/Harvard Medical School, Boston, Massachusetts, USA. 2Harvard Stem Cell Institute, Harvard Medical School, Boston, Massachusetts, USA. 3Erasmus University, Rotterdam, Netherlands. 4Cancer Science Institute, National University of Singapore, Singapore.
Address correspondence to: Daniel G. Tenen, Center for Life Sciences, 3 Blackfan Circle, Room 437, Boston, Massachusetts 02115, USA. Phone: 617.735.2205; Fax: 617.735.2222; E-mail: dtenen@bidmc.harvard.edu.
Authorship note: Hanna S. Radomska and Meritxell Alberich-Jordà contributed equally to this work.
Find articles by Will, B. in: JCI | PubMed | Google Scholar
1Beth Israel Deaconess Medical Center/Harvard Medical School, Boston, Massachusetts, USA. 2Harvard Stem Cell Institute, Harvard Medical School, Boston, Massachusetts, USA. 3Erasmus University, Rotterdam, Netherlands. 4Cancer Science Institute, National University of Singapore, Singapore.
Address correspondence to: Daniel G. Tenen, Center for Life Sciences, 3 Blackfan Circle, Room 437, Boston, Massachusetts 02115, USA. Phone: 617.735.2205; Fax: 617.735.2222; E-mail: dtenen@bidmc.harvard.edu.
Authorship note: Hanna S. Radomska and Meritxell Alberich-Jordà contributed equally to this work.
Find articles by Gonzalez, D. in: JCI | PubMed | Google Scholar
1Beth Israel Deaconess Medical Center/Harvard Medical School, Boston, Massachusetts, USA. 2Harvard Stem Cell Institute, Harvard Medical School, Boston, Massachusetts, USA. 3Erasmus University, Rotterdam, Netherlands. 4Cancer Science Institute, National University of Singapore, Singapore.
Address correspondence to: Daniel G. Tenen, Center for Life Sciences, 3 Blackfan Circle, Room 437, Boston, Massachusetts 02115, USA. Phone: 617.735.2205; Fax: 617.735.2222; E-mail: dtenen@bidmc.harvard.edu.
Authorship note: Hanna S. Radomska and Meritxell Alberich-Jordà contributed equally to this work.
Find articles by Delwel, R. in: JCI | PubMed | Google Scholar
1Beth Israel Deaconess Medical Center/Harvard Medical School, Boston, Massachusetts, USA. 2Harvard Stem Cell Institute, Harvard Medical School, Boston, Massachusetts, USA. 3Erasmus University, Rotterdam, Netherlands. 4Cancer Science Institute, National University of Singapore, Singapore.
Address correspondence to: Daniel G. Tenen, Center for Life Sciences, 3 Blackfan Circle, Room 437, Boston, Massachusetts 02115, USA. Phone: 617.735.2205; Fax: 617.735.2222; E-mail: dtenen@bidmc.harvard.edu.
Authorship note: Hanna S. Radomska and Meritxell Alberich-Jordà contributed equally to this work.
Find articles by Tenen, D. in: JCI | PubMed | Google Scholar
Authorship note: Hanna S. Radomska and Meritxell Alberich-Jordà contributed equally to this work.
Published July 17, 2012 - More info
J Clin Invest. 2012;122(8):2955–2966. https://doi.org/10.1172/JCI43354.
© 2012 The American Society for Clinical Investigation
Received: April 15, 2010; Accepted: June 7, 2012
-
Abstract
Mutations that activate the fms-like tyrosine kinase 3 (FLT3) receptor are among the most prevalent mutations in acute myeloid leukemias. The oncogenic role of FLT3 mutants has been attributed to the abnormal activation of several downstream signaling pathways, such as STAT3, STAT5, ERK1/2, and AKT. Here, we discovered that the cyclin-dependent kinase 1 (CDK1) pathway is also affected by internal tandem duplication mutations in FLT3. Moreover, we also identified C/EBPα, a granulopoiesis-promoting transcription factor, as a substrate for CDK1. We further demonstrated that CDK1 phosphorylates C/EBPα on serine 21, which inhibits its differentiation-inducing function. Importantly, we found that inhibition of CDK1 activity relieves the differentiation block in cell lines with mutated FLT3 as well as in primary patient–derived peripheral blood samples. Clinical trials with CDK1 inhibitors are currently under way for various malignancies. Our data strongly suggest that targeting the CDK1 pathway might be applied in the treatment of FLT3ITD mutant leukemias, especially those resistant to FLT3 inhibitor therapies.
-
Introduction
In acute myeloid leukemia (AML), an immature cell can acquire genetic changes, such as chromosomal translocations, insertions, deletions, or point mutations, which lead to uncontrolled cell growth, protection against cell death, and differentiation arrest. Among the most common oncogenic mutations in AML are internal tandem duplications (ITD) or activating mutations in fms-like tyrosine kinase 3 (FLT3). FLT3 is normally expressed in early hematopoietic precursors and plays a role in their proliferation and differentiation (1, 2), but its aberrant activation contributes to the development of AML. FLT3ITD mutations occur in about 20%–30% of AML patients, and the majority of these mutations (over 70%) are located in the juxtamembrane domain of FLT3. A novel type of ITD mutation (over 28%) was recently identified within the first kinase domain of the receptor (3). Several amino acids in the kinase domain are also known to undergo activating point mutations, for example, mutations in aspartic acid 835, which are seen in about 7% of AML cases (4). The consequences of FLT3 mutations are self phosphorylation and ligand-independent activation of the FLT3 receptor, followed by activation of the downstream signaling pathways, mainly Stat5, Akt, ERK1/2, Pim-1/2, and SHP-1 (5–11). Patients with activating FLT3 mutations have a poor prognosis (1, 2, 4, 12–14); therefore, much effort is being put forth to develop specific therapies. Small molecule inhibitors that specifically inhibit the FLT3 activity are presently undergoing clinical trials (1, 2, 4, 12–16). We have previously demonstrated that one of the targets of the ERK1/2 kinase is C/EBPα, a transcription factor playing a critical role in granulocytic differentiation (17) and often inactivated in various subtypes of leukemia by multiple mechanisms, such as transcriptional and translational silencing, as well as genetic mutations and posttranslational modifications, which render C/EBPα protein nonfunctional. The importance of C/EBPα as a molecular switch is underscored by the fact that it is both necessary and sufficient for granulocytic differentiation (18, 19). Activity of C/EBPα can be modulated by phosphorylation, and a number of residues in the C/EBPα protein that are subject to modifications have been identified. However, until now, only phosphorylation of serine 21 has been shown to have clinical importance (20, 21). We have shown that this single amino acid modification by the ERK1/2 pathway inhibits the function of C/EBPα and is responsible for the differentiation block in FLT3ITD leukemic blasts (17, 21). Pharmacological or genetic abrogation of this phosphorylation event in leukemic cells, for example, treatment with MEK1 inhibitor or substitution with a nonphosphorylatable mutant of C/EBPα (S21A), permits granulopoiesis to proceed (17, 21). Phosphorylation of C/EBPα on serine 21 by p38 MAPK in hepatocytes, on the other hand, increases its transactivation potential on the phosphoenolpyruvate carboxykinase (PEPCK) gene promoter and results in increased PEPCK expression (20). Thus, serine 21 phosphorylation in liver enhances gluconeogenesis and, therefore, may play a role in diabetes.
Interestingly, among FLT3ITD patients, only 39% demonstrated activation of MEK1, and thus the ERK1/2 pathway (22), yet C/EBPα can still be inactivated by phosphorylation on serine 21 (this study). Herein, we identified cyclin-dependent kinase 1 (CDK1, also known as CDC2) as an FLT3ITD-activated kinase, which is responsible for C/EBPα phosphorylation on serine 21 and the blocking of its function. Thus, we provide a molecular mechanism by which the constitutively active FLT3 mutant receptor contributes to the pathogenesis of leukemia, and we propose the use of CDK1 inhibitors for the treatment of FLT3ITD leukemia.
-
Results
C/EBPα transcription factor can be phosphorylated on serine 21 by an ERK1/2-independent kinase. We reported previously that the granulocytic differentiation-promoting function of C/EBPα could be inhibited in FLT3ITD AML by ERK1/2-mediated phosphorylation of serine 21 (17, 21). It has also been reported that not every FLT3ITD mutant can constitutively activate the ERK1/2 pathway (22, 23). To test whether the differentiation block in FLT3ITD AML can be mediated by an ERK-independent phosphorylation of C/EBPα, we transiently coexpressed C/EBPα and FLT3ITD mutant N51 (24), known not to activate the ERK1/2 pathway (ref. 23 and Radomska, unpublished observations), or empty vector (MSCV-IRES-EGFP) in 293T cells and analyzed phosphorylation of C/EBPα on serine 21 by Western blot. Figure 1 shows that while activation of the ERK1/2 pathway by brief treatment with TPA in the absence of FLT3ITD N51 resulted in a subtle increase in phosphorylation of serine 21, a robust phosphorylation took place when C/EBPα was coexpressed with FLT3ITD N51. Consistent with the previous report (23), overexpression of FLT3ITD N51 in the absence of TPA stimulation did not activate the ERK1/2 pathway, and the treatment with MEK1/2 inhibitor PD98059 did not affect the serine 21 phosphorylation levels. In contrast, treatments with FLT3 inhibitors MLN518 and PKC412 led to a substantial decrease in serine 21 phosphorylation. These results strongly suggest that FLT3ITD mutant(s) can activate a novel pathway other than ERK1/2, which is capable of phosphorylating C/EBPα on serine 21 and blocking its function.
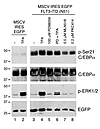 Figure 1
Figure 1FLT3ITD can induce phosphorylation of C/EBPα on serine 21 by a non-ERK1/2 pathway. 293T cells were transiently cotransfected with a C/EBPα expression vector together with an empty MSCV-IRES-EGFP vector (lanes 1 and 2) or MSCV-IRES-EGFP–expressing FLT3ITD mutant N51 (lanes 3–8). Whole-cell extracts were analyzed by Western blot. The same membrane was stained sequentially with antibodies indicated to the right. Samples shown in lanes 1 and 3 were left untreated. Cells were also treated with TPA for 15 minutes to activate the ERK1/2 pathway (lanes 2 and 4) in the absence or presence (lane 6) of the MEK1/2 inhibitor PD98059. Lanes 7 and 8 contain samples of cells treated with FLT3 inhibitors, MLN518 and PKC412, respectively. All samples shown were analyzed on the same blot.
Phosphorylation of C/EBPα on serine 21 by CDK1 in vitro and in vivo. To identify which kinase, in addition to ERK1/2, can phosphorylate serine 21, we analyzed the amino acid sequence of C/EBPα in the vicinity of serine 21 using Scansite software ( http://scansite.mit.edu/) and found that this residue lies within a motif likely to be phosphorylated by CDK1. To verify this, we performed cell-free in vitro kinase assay using purified active CDK1 kinase and GST-C/EBPα proteins as substrates. As demonstrated in Figure 2A, 32P was specifically incorporated into the GST-C/EBPα WT protein only in the presence of the active enzyme, and mutating serine 21 to alanine abolished phosphorylation. Phenylalanine 31 is located within the docking site for ERK1/2, and it was demonstrated to be necessary for substrate recognition and phosphorylation of C/EBPα on serine 21 (17). In contrast to ERK1/2-mediated phosphorylation, mutating this residue to alanine had no effect on phosphorylation of C/EBPα by CDK1 (Figure 2A).
 Figure 2
Figure 2C/EBPα phosphorylation on serine 21 by CDK1 in vitro and in vivo. (A) In vitro kinase assay. Active CDK1 enzyme was incubated with GST (lane 1) or GST-C/EBPα fusion proteins (lanes 2–5) in the presence of 32P. Lanes 2 and 3 contain WT C/EBPα-GST as a substrate. Lane 4 contains Ser21Ala mutant, and lane 5 contains Phe31Ala mutant. Proteins were resolved on acrylamide gel and blotted to nitrocellulose membrane. Upper panel: autoradiograph of the membrane. Lower panel: staining with the N-terminal C/EBPα antibody. (B) Effect of CDK1 overexpression or mitotic arrest on C/EBPα phosphorylation. 293T cells transfected with empty vector, or HA-tagged CDK1 expression vector (lanes 1 and 2). Cells expressing empty vector (lane 3) or a dominant negative form of CDK1 (lane 4; DN CDK1) were arrested at mitosis by nocodazole treatment. All samples were analyzed on the same gel, and the Western blot was stained with the antibodies indicated. (C) CDK1 silencing results in decreased C/EBPα phosphorylation on serine 21. MOLM-14 cells were transduced with a CDK1 shRNA, sorted for EGFP expression, and cultured for 2 days (total of 4 days of viral transduction). Whole-cell extracts were analyzed by Western blotting with the indicated antibodies. Signals were quantified and graphed. Black bars indicate the relative expression levels of CDK1 and C/EBPα proteins or ERK1/2 activity following the knockdown (compared with white bars for negative controls; NC). Gray bar shows the level of phospho–serine 21 C/EBPα relative to the total C/EBPα protein in cells with downregulated CDK1 expression. sh, short hairpin.
Furthermore, in transiently transfected 293T cells, overexpression of CDK1 led to an increase in serine 21 phosphorylation of cotransfected C/EBPα without changing the activity of ERK1/2 (Figure 2B). It has been demonstrated that during mitosis CDK1 kinase reaches its highest activity level, while ERK1/2 activity subsides (25). Indeed, when C/EBPα-transfected 293T cells were arrested at mitosis by nocodazole treatment, serine 21 was phosphorylated despite the absence of detectable ERK1/2 activity, and this phosphorylation process was abrogated by coexpression of dominant negative mutant of CDK1 (DN CDK1; Figure 2B). Mitotic arrest of U937 cells with nocodazole also showed increased phosphorylation of endogenous C/EBPα, which decreased upon release from arrest (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI43354DS1). Consistent with CDK1-mediated phosphorylation of C/EBPα, treatment of mitotic U937 cells with MEK1 or CDK2/CDK5 inhibitors had no effect on phosphorylation of serine 21, while inhibition of CDK1 did (Supplemental Figure 1B). To further prove the direct role of CDK1 in phosphorylating C/EBPα on serine 21, we performed a knockdown experiment. shRNA specifically targeting CDK1 was expressed from a retroviral vector in MOLM-14 cells. Following sorting of GFP+ cells, the effectiveness of CDK1 knockdown and its effect on phosphorylation of C/EBPα were measured by Western blot. As shown in Figure 2C, normalization for the β-actin levels demonstrated that CDK1 protein expression was decreased by about 60%. There was no change in C/EBPα total protein expression, but there was an approximately 50% decrease in phosphoserine 21 containing C/EBPα species. Notably, CDK1 knockdown had no effect on the levels of active ERK1/2 kinase. Taken together, our results identify serine 21 of C/EBPα as a substrate for CDK1 kinase.
CDK1 pathway is activated in FLT3ITD AML. Constitutively active FLT3ITD receptor kinase has been shown to stimulate a number of downstream pathways. To determine whether CDK1 can be superactivated by FLT3ITD as well, the CDK1 kinase complexes were immunoprecipitated from asynchronously growing FLT3ITD AML cell lines and used in in vitro kinase reaction with histone H1 as a substrate. As shown in Figure 3A, all untreated and DMSO-treated cell lines exhibited high CDK1 activity. In contrast, the kinase activity was significantly repressed by the treatment with the FLT3 inhibitor MLN518 (Figure 3A). To ascertain whether the effect of FLT3ITD on CDK1 is direct or indirect, we examined the cell-cycle distribution of MOLM-14 cells treated with MLN518 or DMSO. Figure 3B shows that FLT3 inhibitor treatment led to a significant decrease in mitotic cells with enrichment of G0-arrested cells. Taken together, these data indicate that CDK1 is a downstream pathway activated by FLT3ITD mutant receptors in an indirect fashion.
 Figure 3
Figure 3Inhibition of constitutive activity of FLT3 decreases CDK1 kinase activity in FLT3ITD AML cells and slows down the cell-cycle progression. (A) CDK1 was immunoprecipitated from FLT3ITD-expressing cell lines (MV4;11, MOLM-13, and MOLM-14) and used in kinase reactions with histone H1 as a substrate and 32P. Proteins were separated on acrylamide gels and blotted to nitrocellulose membranes. The top panel shows the autoradiograph of the blot, and the bottom panel shows staining with anti-CDK1 antibody to assure that equal amounts of CDK1 were immunoprecipitated. The assay was done either in the absence of treatment (–; lanes 1, 4, and 7), after 24-hour treatment with 0.1% DMSO (DMSO; lanes 2, 5, and 8), or after 24-hour treatment with FLT3 inhibitor (MLN518; lanes 3, 6, and 9). The percentages indicate the CDK1 activities remaining after FLT3 inhibitor treatment (DMSO-treated samples were set to 100%). (B) Cell-cycle distribution of MOLM-14 cells treated with DMSO (left panel) or FLT3 inhibitor MLN518 (right panel). Inhibition of FLT3 arrests majority of MOLM-14 cells at G0. Numbers indicate the percentage of cells in each quadrant.
Pharmacological and genetic inhibition of CDK1 activity in FLT3ITD AML relieves differentiation block. To determine the biological effect of CDK1 inhibition in FLT3ITD AML cells, we cultured MV4;11, MOLM-13, and MOLM-14 cells with small molecule inhibitors targeting CDK1: flavopiridol, roscovitine (both being presently tested in clinical trials; http://clinicaltrials.gov), and NU6102. For comparison, we also treated these cells with the FLT3 inhibitor MLN518, which we previously demonstrated as decreasing ERK1/2 activity and phosphorylation of C/EBPα (21), and herein we showed that it can also inhibit CDK1 activity (Figure 3). Figure 4A shows the Western blot results obtained for MOLM-14 cells; comparable results were found for MOLM-13 and MV4;11 cells (data not shown). The treatments with all CDK1 inhibitors tested as briefly as 18 hours resulted in substantial hypophosphorylation of C/EBPα. While exposure to 10 μM NU6102 decreased the levels of phosphoserine 21-C/EBPα by 40%–60% in all 3 cell lines (Figure 4A and data not shown), a decrease in serine 21 phosphorylation by 53%–86% was achieved with 100 nM flavopiridol (Figure 4A and data not shown) and 77%–89% by 25 μM roscovitine (Figure 4A and data not shown). Neither of these CDK1 inhibitors downmodulated the ERK1/2 activity (Figure 4A). In addition to affecting CDK1 activity, all 3 compounds are also known to exert inhibitory effect against a kinase highly homologous to CDK1, CDK2. To eliminate the involvement of CDK2 in phosphorylation of C/EBPα, we cultured the cells in the presence of another compound, PNU 112455A, which inhibits CDK2 and CDK5 (IC50 = 2 μM for both) but does not display activity against other kinases at concentrations as high as 100 μM. In contrast with NU6102, PNU 112455A had no effect on phosphorylation of C/EBPα in all cells (Figure 4A and data not shown).
 Figure 4
Figure 4Granulocytic differentiation of FLT3ITD cells after CDK1 inhibition. (A) Inhibition of CDK1 but not CDK2/CDK5 leads to hypophosphorylation of C/EBPα on serine 21. MOLM-14 cells were treated with CDK1 inhibitors NU6102 (lane 3), flavopiridol (flavo; lanes 6–7), or roscovitine (rosco; lanes 8–9) for 18 hours at the concentrations indicated. For control, cells were treated with DMSO (lanes 1 and 5). Treatment with the FLT3 inhibitor MLN518 (lane 2) was used for a positive control. Lane 4 shows cells cultured in the presence of CDK2/CDK5 inhibitor PNU 112455A (PNU). Shown is a Western blot stained with antibodies indicated. (B) Morphological differentiation of MOLM-14 cells treated with CDK1 inhibitors. Cells were either untreated or treated with 0.1% DMSO for 3 days (DMSO), 10 mM PNU112455A for 3 days (PNU112455A), 10 mM NU6102 for 3 days (NU6102), 12.5 mM roscovitine for 2 days, or 100 nM flavopiridol for 2 days (flavo). Cytospin was stained with Wright-Giemsa. Red arrowheads point to the cells displaying granulocytic maturation. Original magnification, ×40. (C) Downregulation of c-myc expression in response to CDK1 inhibition. MOLM-14 cells were grown in the presence of the indicated drugs for 18 hours and analyzed by Western blot. (D) Increase in CD11b surface expression on MOLM-14 cells treated with CDK1 inhibitor. Cells were treated with CDK1 inhibitor NU6102 or vehicle control (0.1% DMSO) for up to 4 days and analyzed daily for the expression of CD11b by flow cytometry. Left panel: histograms obtained on day 3. Right panel: y axis indicates the percentages of CD11b+ cells during the time course.
We have previously reported that C/EBPα, when hypophosphorylated on serine 21, displays granulocytic differentiation-promoting activity (17, 21). We cultured FLT3ITD cell lines in the presence of CDK1 inhibitors for up to 3 days and monitored their morphology as well as the changes in maturation marker expression. As shown in Figure 4B, MOLM-14 cells treated with 10 μM NU6102 acquired granulocytic morphology as early as on day 2 of the culture, with more marked effect seen on day 3. Similar changes in cell morphology were also noted after 2 days of treatment with 12.5 μM roscovitine or 100 nM flavopiridol, although a rapid onset of apoptosis was more pronounced in those cultures (Figure 4B and data not shown). No morphological changes were observed when the cells were treated with a vehicle control, DMSO, or CDK2/CDK5 inhibitor PNU 112455A (Figure 4B). Comparable results were obtained for MOLM-13 cells (data not shown), while for MV4;11 cells, morphological changes were less pronounced (data not shown) and not seen with non-FLT3ITD AML cells (U937, KG1a, and K562; data not shown). All 3 FLT3ITD cell lines treated with NU6102 also demonstrated downregulation of c-myc, which is indicative of myeloid maturation (Figure 4C and data not shown). In addition, NU6102 treatment led to a time-dependent increase in the number of surface CD11b–expressing MOLM-14 cells (up to 60% on days 3 and 4; Figure 4D), which is in accord with granulocytic differentiation. The ERK1/2 pathway was originally discovered to be responsible for phosphorylation of serine 21 (17, 21). Previously, we reported that inhibition of this pathway in FLT3ITD-expressing MV4;11 cells decreased phosphorylation of C/EBPα and induced granulocytic differentiation (21). In MOLM-14 cells, inhibition of the ERK1/2 pathway also led to differentiation with similar kinetics, but the effect of the inhibition of CDK1 was more potent, as measured by the downregulation of c-myc, upregulation of CD11b surface expression, increase in myeloperoxidase (MPO) and lysozyme mRNA expression, and morphological changes (Supplemental Figure 2).
Next, we determined whether the differentiation-promoting effect of CDK1 inhibitors was dependent on C/EBPα expression. We designed an shRNA lentiviral construct specifically targeting CEBPA and demonstrated C/EBPα downregulation at the protein level (Supplemental Figure 3). As expected, MOLM-14 cells transduced with a nonsilencing control shRNA showed upregulation of CD11b expression upon treatment with the CDK1 inhibitor NU6102 in comparison with the DMSO-treated cells, whereas MOLM-14 cells transduced with the C/EBPα shRNA did not respond to the treatment (Figure 5A). These changes observed by flow cytometry nicely correlated with the morphological analysis of cytospun cells (Figure 5B). These data indicate that the differentiation-inducing effects of CDK1 inhibitors are C/EBPα dependent.
 Figure 5
Figure 5Downregulation of C/EBPα expression prevents the granulocytic differentiation induced by CDK1 inhibitors. (A) MOLM-14 cells were transduced with a nonsilencing control shRNA (NSC) or an shRNA-targeting C/EBPα (C/EBPα sh) lentivirus. Cells were treated with either DMSO control (D) or 5 mM NU6102 (NU). The y axis indicates the percentage of CD11b-positive cells after 4 days of culture. (B) Morphological analysis of MOLM-14 cells upon C/EBPα silencing and CDK1 inhibitor treatment. Cells infected with the NSC or the C/EBPα shRNA lentivirus were treated for 4 days in the presence of 5 mM NU6102. Cytospins were stained with Wright-Giemsa. Original magnification, ×40.
To test whether specific knockdown of CDK1 protein expression would have the same effect as treatments with small molecule compounds, MOLM-14 cells were transduced with viral particles expressing CDK1 shRNA and EGFP. We anticipated that rapid inhibition of CDK1, which is necessary for cell-cycle progression through mitosis, may lead to growth arrest and apoptosis. We assumed that the expression level of CDK1 shRNA might parallel the level of EGFP expression and thus be in inverse correlation with the expression of the endogenous CDK1 protein. In order to provide the gradient of CDK1 knockdown, EGFP+ cells were sorted into 3 populations with low, medium, and high EGFP intensities. Each population was maintained in complete culture medium, and cell morphology was monitored daily. On day 4, we harvested 30,000 cells from each population, made lysates, and analyzed the degree of knockdown by Western blot. Day 4 was selected based on our previous knockdown experiment, showing that this was the earliest time point demonstrating detectable and significant decrease in CDK1 protein expression. Figure 6A shows that after normalization for the β-actin protein, there was not much difference between the inhibition of CDK1 protein expression in cells expressing medium and high levels of EGFP. Cells with low EGFP had a modest, but detectable decrease in CDK1. Morphological examination showed that cells sorted for high EGFP were enlarged in size, but did not show clear signs of myeloid differentiation. They also became growth arrested and died on day 7 (data not shown). Cells sorted for medium and low EGFP levels, on the other hand, acquired morphological changes consistent with granulocytic maturation on day 10 (Figure 6B), and this effect was stronger for low EGFP–expressing cells. Medium EGFP–expressing cells, in addition to myeloid maturation, were accompanied by severe cell death (data not shown).
 Figure 6
Figure 6Induction of granulocytic differentiation following genetic downregulation of CDK1 expression. (A) shRNA-mediated decreased expression of CDK1 in MOLM-14. MOLM-14 cells were virally transduced with CDK1-targetting shRNA (CDK1 sh) or negative control. Two days later, populations expressing low (Lo), medium (Med), or high (Hi) GFP levels were sorted. Following an additional 2 days of culture, an aliquot of each cell population was lysed and analyzed by Western blot with anti-CDK1 antibody (top panel). β-actin antibody was used for loading control. All samples were loaded on the same gel but were noncontiguous. Quantifications of the band intensities are shown below. Neg, negative for GFP. (B) Downregulation of CDK1 expression leads to granulocytic maturation. Following sorting, cells were cultured for up to 8 more days and their morphology was monitored daily on Wright-Giemsa–stained slides. Original magnification, ×40.
Finally, primary FLT3ITD leukemic samples collected at diagnosis from the peripheral blood of patients were treated with NU6102. As expected, CDK1 inhibition led to a remarkable hypophosphorylation of C/EBPα in the FLT3ITD patient cells in 3 out of 4 FLT3ITD patient samples (Figure 7). Figure 7 shows the Western blot analysis after 24 hours of treatment, although the effect was already noticeable after 10 hours. Phosphorylation of serine 21 was also observed in leukemic samples with the WT FLT3 gene (Figure 7), most likely due to constitutive activation of the ERK pathway. This is in agreement with the findings that the enhanced activity of this pathway was detected in over 80% of the AML samples tested, regardless of their FLT3 genotype status (26, 27). Nevertheless, the samples not harboring FLT3ITD mutations showed negligible or no effect upon treatment with the CDK1 inhibitor (Figure 7). In addition, 8 patient samples carrying FLT3ITD and 2 patient samples with the WT FLT3 receptor were also examined for the expression of mature (CD11b, CD11c, CD14, G-CSF-R, CD15, and CD16) and immature (CD33, CD34, CD38, and CD133) cell-surface markers following treatment with NU6102. Although each sample demonstrated different specific profiles of the response, all patient samples carrying FLT3ITD showed a general tendency to increase the expression of maturation markers and decrease that of immature markers (Figure 8A). In contrast, samples with WT FLT3 (patients C and D), which did not show a decrease in serine 21 phosphorylation, did not show any signs of differentiation (Supplemental Figure 4). Whenever we had enough material (patients A, F, and G), we also tested the mRNA expression of granulocyte-specific genes, such as CEBPE, CSF3R (coding for G-CSF receptor), neutrophil elastase, gelatinase A, and lysozyme. All 3 patient samples showed increases in CSF3R and CEBPE expression after treatment with CDK1 inhibitor; patient F demonstrated an increase in expression of all 5 genes (Figure 8B). Moreover, the treatment of FLT3ITD-carrying specimens with NU6102 for 7 days was accompanied by morphological changes, suggesting granulocytic differentiation (Supplemental Figure 5). Since CDK1 inhibition was associated with substantial cell death, we wanted to make sure that lobing of the nuclei seen on cytospin preparations was caused by true differentiation, rather than apoptosis. Therefore, FLT3ITD leukemic samples treated with NU6102 were sorted for CD15+ cells and tested by the Wright-Giemsa method. As shown in Figure 8C, the same morphological forms were observed and importantly, CD15+ cells were nearly 90% viable (Figure 8C). In summary, inhibition of CDK1 in FLT3ITD cells led to an increase in C/EBPα function by its hypophosphorylation on serine 21 and the relief of the differentiation arrest.
 Figure 7
Figure 7Decreased C/EBPα phosphorylation upon CDK1 inhibition in FLT3ITD AML patients. Four patient samples with FLT3ITD mutations (patients A, B, F, and H) and 4 patient samples with WT FLT3 receptor (patients C, D, K, and L) were cultured in vitro for 24 hours in the presence of 10 mM NU6102 or 0.1% DMSO. Whole-cell lysates were tested by Western blot. Signals for serine 21–phosphorylated proteins were normalized for the total C/EBPα protein and plotted (shown below Western blot; D, DMSO, N, NU6102). The numbers above bars indicate the percentage of phosphorylated C/EBPα species compared with samples treated with DMSO (set to 100%).
 Figure 8
Figure 8Granulocytic differentiation of FLT3ITD cells after inhibition of CDK1 activity. (A) Inhibition of CDK1 in patient samples with FLT3ITD induces granulocytic differentiation. Blood specimens from FLT3ITD AML patients (patient A and patient B; same as shown in Figure 7) were cultured in the presence of 0.1% DMSO or 10 mM NU6102 for 7 days. Cell aliquots were stained with anti-CD11c, anti-CD15, anti-CD14, anti-CD11b, anti-CD16, anti–G-CSF-R, anti-CD33, anti-CD133, anti-CD34, and anti-CD38 antibodies and analyzed by flow cytometry. The y axes indicate the percentage of positive cells. (B) Patient samples were treated as in A for 5 days and analyzed for mRNA expression of granulocytic cell-surface markers by quantitative RT-PCR. The y axes indicate relative expression to GAPDH. (C) CD15+ cells with granulocytic-like morphology are viable. CD15 expression on FLT3ITD AML patient A sample treated with DMSO (gray) or 5 mM NU6102 (white) during 10 days (left panel). Number indicates the percentage of CD15+ cells upon NU6102 treatment. CD15+ cells were sorted, cytocentrifuged, and stained with Wright-Giemsa method (middle panel). Original magnification, ×40. CD15+ cells were also stained with annexin V and PI to determine the extent of their viability (right panel). The numbers in each quadrant indicate percentages of viable (lower left), early apoptotic (lower right), late apoptotic (upper right), and necrotic cells (upper left).
Activity of CDK1 is controlled by a multiple-step process, but ultimately, CDK1 can be activated by binding to cyclin B1 (28, 29). To determine how the constitutively active FLT3ITD receptor affects CDK1 activity, MOLM-14 cells were untreated or treated with FLT3 inhibitor MLN518 or vehicle control DMSO and analyzed by Western blot staining with anti–cyclin B1 antibody. As shown in Figure 9A, treatment of MOLM-14 cells with FLT3 inhibitor led to a decreased expression of cyclin B1 protein. Conversely, introduction of FLT3ITD into Ba/F3 cells led to about a 2-fold increase in total cyclin B1 protein levels (Figure 9B). These data suggest a possible involvement of cyclin B1 in the activation of CDK1.
 Figure 9
Figure 9The activity of CDK1 in FLT3ITD-expressing MOLM-14 cells is correlated with cyclin B1 protein levels. MOLM-14 cells were left untreated (U) or treated for 24 hours with either 0.05% DMSO or 5 mM MLN518 FLT3 inhibitor (MLN), and whole-cell lysates were analyzed by Western blot with antibodies indicated on the right. (A) Cyclin B1 protein levels decrease upon inhibition of FLT3 receptor in MOLM-14 cells. (B) Forced expression of FLT3ITD mutant (N51) in BaF3 cells leads to 2.1-fold increase in cyclin B1.
-
Discussion
Activating mutations in FLT3 receptor tyrosine kinase are among the most common mutations in AML and indicate poor prognoses (1, 2, 4, 12–14). Thus, development of small molecule inhibitors specifically targeting FLT3 seemed to be a promising approach for treating a large number of AML cases (15). Although different activating mutations in FLT3 exhibit divergent sensitivities toward different FLT3 inhibitors (30), the preclinical and clinical trials involving the first generation of drugs (PKC-412, MLN518, CEP-701, SU11248, and AC220) showed biologic activity and favorable toxicity (15, 16, 31–36). However, in several cases, patients demonstrated primary resistance to the drugs (3, 37). Alternatively, an initial response was soon followed by emergence of secondary mutations, for example, mutations N676K and D835Y in the kinase domain of FLT3 (38, 39). To bypass this problem, a search for a new generation of FLT3 inhibitors is under way. In the meantime, combination therapies with selective FLT3 inhibitors and conventional cytotoxic chemotherapy are being investigated, but those have been characterized by unacceptable toxicity and poor tolerance (40). Alternative treatments of FLT3 mutant AML may involve inhibitors of the downstream pathways activated by FLT3ITD mutations. For example, we demonstrated before that inhibition of the ERK1/2 pathway in FLT3ITD-expressing AML cells leads to their differentiation (21). The multitude of downstream signaling pathways activated by mutant FLT3 receptors (5–11) may increase the repertoire of possible drug combinations.
A majority of the FLT3ITD downstream pathways control survival and apoptosis, while ERK1/2 signaling plays a role in the differentiation block by phosphorylating the C/EBPα transcription factor on serine 21 and inhibiting its function (17, 21). Interestingly, only a fraction of FLT3ITD patients exhibited activation of the ERK1/2 pathway (22). Of note, due to technical difficulties in examining the ERK activity in FLT3ITD leukemias, additional studies are needed to determine whether activation of ERK can serve as a specific biomarker of FLT3 signaling in primary leukemias (22). Moreover, we observed serine 21 phosphorylation on C/EBPα in cells with an FLT3ITD mutant receptor, which is disabled in ERK1/2 activation (mutant N51; refs. 23, 24). We hypothesized that a kinase other than ERK1/2 may be responsible for C/EBPα phosphorylation and differentiation block. In this report, we identified CDK1 (also known as CDC2) as the kinase specifically modifying C/EBPα on serine 21 in AML with FLT3ITD mutations. Thus, our data provide a potential molecular mechanism explaining the maturation block in FLT3ITD cases without activation of ERK1/2. However, in addition to ERK1/2, CDK1 is another modulator of C/EBPα differentiation function, and we cannot discard the contribution of other mediators to the differentiation block seen in FLT3ITD AML. Further, our observations do not rule out a potential interplay between ERK1/2 and CDK1 activity on C/EBPα function in certain FLT3ITD AML cases. The FLT3ITD and CDK1 connection was previously reported by Odgerel et al. (41). While they reported that CDK1 is partially inactivated in FLT3ITD AML cell lines, our work concludes that CDK1 can be activated by FLT3ITD mutations. This apparent contradiction could be explained by the use of different FLT3 inhibitors (PKC412 and MLN518, respectively) and the concentrations used, resulting in either effects in apoptosis (41) or differentiation (this study).
Several studies described modulation of C/EBPα activity by phosphorylation on various residues (17, 42–44). However, phosphorylation of a single amino acid, serine 21, seems to have the most remarkable effect by shifting the activity of C/EBPα from a granulocytic differentiation-promoting factor (unphosphorylated) to a dominant negative form (phosphorylated) (17). Serine 21 can be phosphorylated by ERK1/2 (17, 21), p38 MAPK (20, 45), and CDK1 (this report). It is the only phosphorylation site on C/EBPα with clinical significance identified so far. Hyperphosphorylation of serine 21 in leukemic blasts blocks their maturation (21). P38MAPK-mediated phosphorylation of serine 21-C/EBPα acts as a switch inhibiting neutrophilic differentiation of CD34+ progenitors while permitting their eosinophilic maturation and may be responsible for disturbed neutrophilic development in severe congenital neutropenia (45). In liver cells, however, phosphorylation of serine 21 by p38 MAPK increases the activity of C/EBPα on promoters of genes involved in gluconeogenesis, thus possibly contributing to diabetes (20, 46).
CDK1 belongs to a family of cyclin-dependent kinases, which are critical regulators of cell division. While individual CDK proteins may substitute for each other’s function, gene-targeting experiments demonstrated that CDK1 is the only family member whose role in promoting mitosis cannot be substituted by any other CDK (47). Misregulation of CDK1 expression/activity in solid tumors is well documented, and several clinical trials with CDK1 inhibitors are currently under way. In contrast, there is very little known about the role of CDK1 during leukemogenesis. Higher expression levels of CDK1 were detected in leukemic cells with del(5q) (48). Also, a recent report described upregulation of CDK1 in leukemia with translocation liposarcoma/ETS-related gene (TLS-ERG) and attributed increased expression of CDK1 to the differentiation block (49). Similarly, we found that in AML with constitutively active FLT3 receptor, CDK1 can contribute to the maturation block by inhibiting function of the transcription factor C/EBPα, which is required for granulopoietic development. Our findings open the door to the possibilities of using pharmacological inhibitors of CDK1 in leukemia as well.
Studies described herein demonstrated that various CDK1 inhibitors affect C/EBPα phosphorylation and promote the differentiation process to variable degrees. This might be due to their broad spectrum of action; by inhibiting multiple pathways at the same time, they can rapidly induce apoptosis. While we believe that much of the activity of these inhibitors is through their effects on C/EBPα phosphorylation, our experiments do not rule out effects on other pathways as well. We also found that activation of the CDK1 pathway by FLT3ITD involves upregulation of cyclin B1 rather than upregulation of CDK1 itself (49). While these data suggest that cyclin B1 is involved in the activation of CDK1, we cannot rule out that other CDK1 regulators, such as Myt1 and cdc25c, could also be involved (41). NU6102 demonstrated the best differentiation-promoting activity, perhaps because of the higher specificity against CDK1 versus other CDKs. The knockdown experiments are in accord with this hypothesis. Cells expressing lower levels of EGFP and presumably lower levels of shRNA showed more pronounced maturation, while the cells with higher levels of EGFP (and presumably the highest levels of shRNA) exhibited mainly apoptosis (this study).
In summary, we demonstrate that constitutive activation of the FLT3 receptor can lead to abnormal activation of multiple downstream signaling pathways (Figure 10), which are capable of inhibiting the function of C/EBPα, contributing to the differentiation block. Inhibiting either FLT3 receptor, MEK1 kinase, or CDK1 can restore the activity of C/EBPα and induce myeloid maturation of leukemic blasts.
 Figure 10
Figure 10Effect of constitutive activation of the FLT3 receptor in leukemogenesis. Activation of FLT3 receptor by ITD mutations (stars) promotes cell survival and inhibits apoptosis by activation of STAT3, STAT5, and AKT. Differentiation of myeloid precursors is mediated by the C/EBPα transcription factor, which exhibits full maturation-promoting activity when hypophosphorylated on serine 21. Activation of ERK1/2 and/or CDK1 (via increased expression of cyclin B [Cycl B]) leads to hyperphosphorylation of C/EBPα, which abolishes its function and results in a differentiation block. Broken arrows indicate a likely cascade of downstream pathways from the activated FLT3 leading to upregulation of c-myc and cyclin B. Pharmacological inhibition of either the FLT3, MEK1,or CDK1 pathway results in decreased phosphorylation of C/EBPα on serine 21, increase in its activity, and induction of granulopoiesis.
-
Methods
Cell lines. Human AML lines carrying FLT3ITD mutations were kindly donated by Yoshinobu Matsuo (MOLM-13, Fujisaki Cell Center, Hayashibara Biochemical Labs, and the Kurashiki Medical Center, Kurashiki, Okayama, Japan), Neill Giese (MOLM-14; Calistoga Pharmaceuticals, Inc.), Stefan Heinrichs (MV4;11, Department of Pediatric Oncology, Farber Cancer Institute, Boston, Massachusetts, USA), and David Sternberg (Ba/F3-FLT3ITD, OSI Pharmaceuticals). MOLM-13, MOLM-14 (50), and MV4;11 (CRL 9591; ATCC) as well as U937 (CRL 1593; ATCC) were grown in RPMI 1640 with 10% FBS. The murine bone marrow–derived IL-3–dependent Ba/F3 cell line (51) was cultured in RPMI with 10% FBS and 10% WEHI-3B conditioned medium. A Ba/F3 stable line expressing the FLT3ITD mutant N51 (24) was maintained in RPMI/10% FBS/10% WEHI-3B conditioned medium and 750 μg/ml active G418. Human Embryonal Carcinoma 293T (HEK 293T; CRL 11268, ATCC) and Phoenix-Ampho Packaging (Orbigen) cell lines were cultured in DMEM with 10% FBS.
Patient samples. After informed consent was obtained, peripheral blood samples of AML patients were collected at the time of diagnosis before initiation of treatment. Blasts and mononuclear cells were purified by Ficoll-Hypaque (Nygaard) centrifugation and cryopreserved. Cells were thawed at 37°C, incubated on ice for 10 minutes, and washed twice in ice-cold HBSS. Cells were precultured in EX-VIVO (BioWhittaker) medium supplemented with 10 ng/ml human IL-3 (hIL-3), 10 ng/ml hIL-6, and 25 ng/ml hSCF at 37°C for 45 minutes on 150 mm Petri dishes to remove adherent cells. Suspension cells were collected and incubated at 37°C in the presence of 5 or 10 μM NU6102 or 0.1% DMSO (vehicle control).
Reagents. All inhibitors were prepared in DMSO. FLT3 inhibitors MLN518 (CT53518; Millenium Pharmaceuticals) and PKC-412 (Biomol) as well as CDK1 inhibitors NU6102 (Calbiochem/EMD Biosciences) and flavopiridol (Sigma-Aldrich) were reconstituted at 10 mM. Roscovitine was reconstituted at 25 mM. The CDK2/CDK5 inhibitor PNU112455A (Calbiochem) was dissolved at 10 mM, and the MEK1 inhibitor PD98059 was reconstituted at 50 mM. TPA stock solution was prepared at a 1-mM concentration. All reagents except MLN518 (kept at 4°C) were stored at –20°C. A stock solution of nocodazole (Sigma-Aldrich) was prepared in DMSO at 10 mg/ml.
Plasmids. MSCV-IRES-EGFP-FLT3ITD (N51) was described previously (24). The GST-C/EBPα fusion expression vectors (containing 139 of the N-terminal amino acids of either WT C/EBPα, S21A, or F31A mutations) were described previously (21). WT and dominant negative (DN-CDK1) isoforms of human CDK1 cDNAs were HA tagged at the C termini and cloned in pCMV-neo-Bam expression vector (52). Liu Yang (Departments of Orthopedics and Medicine/Hematology, University of Washington, Seattle, Washington, USA) provided CDK1 siRNA lentiviral constructs. The shRNA oligonucleotide (GGATTCCAGGTTATATCTCATCTCGAGATGAGATATAACCTGGAATCCTTTTTTT; targeting nt 350–370 of the human CDK1 coding sequence) was cloned under the control of the human H1 promoter in pNantx retroviral vector, which contains a GFP reporter (described in ref. 53). The human CEBPA and a nonsilencing control shRNA sequences were cloned into the lentiviral vector pGhU6 containing an EGFP reporter. The shRNA oligonucleotide sequence targeting CEBPA was ACCCCGCCAAGAAGTCGGTGGACAAGAACATCAAGAGTGTTCTTGTCCACCGACTTCTTGGCTTTTTGGAA (930–954 nt), and the nonsilencing control sequence was ACCCCATCTCGCTTGGGCGAGAGTAACATCAAGAGTTACTCTCGCCCAAGCGAGATTTTTTGGAA.
In vitro kinase assays. C/EBPα-GST fusion proteins were isolated from BL21(DE3)pLys cells stimulated with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 1 hour. Bacteria were then centrifuged at 1,500 g for 10 minutes, and bacterial pellets were subjected to a single cycle of freeze-thawing. Lysed bacteria were suspended in 1 ml BugBuster Protein Extraction Agent (Novagen) and supplemented with 25 U of Benzonase Nuclease (Novagen). Following a 5-minute incubation at room temperature and centrifugation at 13,000 g for 20 minutes, the supernatant was collected and incubated with 0.2 ml of Glutathione Sepharose 4FF beads (Amersham) for 1 hour at room temperature. Beads were pelleted and washed 4 times with PBS and immediately suspended in in vitro kinase reaction mixture containing [γ32P]ATP (10 Ci/mmol), purified CDK1 kinase (cat. P6020S; NEB), and supplied reaction buffer. The reactions were carried out at 30°C for 10 minutes, then stopped by adding 4× Laemmli buffer and boiling samples at 100°C for 10 minutes. The products were resolved on SDS-PAGE, blotted to nitrocellulose membrane, and analyzed by autoradiography. To control for the amount of C/EBPα-GST protein, the same membrane was stained with the N-terminal anti-C/EBPα antibody.
In vivo kinase assay. FLT3ITD AML cells were treated with 10 μM MLN518 or 0.1% DMSO for 24 hours. Equal numbers of cells were harvested and lysed in RIPA lysis buffer. Active CDK1 kinase complexes were immunoprecipitated with anti-CDK1 antibody and captured on Immobilized Protein A beads (IPA-300; Repligen). Following 3 washes with ice-cold PBS and 1 wash with ADBI CDK1 kinase reaction buffer (Upstate Biotechnology), the CDK1-containing beads were used in kinase reactions in ADBI containing purified histone H1 as a substrate and [γ32P]ATP (10 Ci/mmol). The reactions were carried out at 30°C for 10 minutes, stopped by boiling in Laemmli buffer for 10 minutes, subjected to SDS-PAGE electrophoresis, and transferred onto nitrocellulose membranes. The incorporation of 32P into the substrate was examined by autoradiography, and the amounts of immunoprecipitated CDK1 kinase were compared by staining the same membrane with anti-CDK1 antibody.
Mitotic arrest. U937 cells were arrested at mitosis by incubation in the presence of 100 μg/ml nocodazole for 16 hours. Cells were released from the block by washing twice and by subsequent culture in complete medium without nocodazole.
Western blot. Typically, 5 × 106 cells were spun (1 K, 5 minutes), washed in PBS, lysed in 400 μl of 1× Laemmli Sample Buffer, and boiled at 100°C for 10 minutes. From 30 to 40 μl of each lysate was loaded on 7.5% SDS-PAGE gels and proteins transferred to nitrocellulose membranes. Following blocking in 5% milk/TBST (TBST: 25 mM Tris-HCl pH 7.4, 137 mM NaCl, 2.7 mM KCl, 0.1% Tween 20), membranes were stained with primary antibodies diluted in 5% BSA/TBST/0.1% sodium azide overnight at 4°C and then with HRP-conjugated secondary antibodies at room temperature for 1 hour. Signals were detected by enhanced chemiluminescence and quantified by ImageQuant software (Molecular Dynamics). The primary antibodies were goat N-terminal C/EBPα (N-19; 1:1,000; sc-9315, Santa Cruz Biotechnology Inc.), rabbit phospho–Ser21-C/EBPα (1:1,000; #2841, Cell Signaling Technology), goat C-terminal C/EBPα (C-18, 1:1,000; sc-9314, Santa Cruz Biotechnology Inc.), rabbit C/EBPα (14AA, 1:1,000; sc-61, Santa Cruz Biotechnology Inc.), rabbit CDK1 (1:1,000; PC25, Calbiochem), rabbit phospho-(T202/Y204)-ERK1/2 (1:1,000; #9101, Cell Signaling Technology), panERK (1: 1,000; #610123, BD Transduction Laboratories), cyclin A (C-19; sc-596) β-actin (1:10,000; A5441, Sigma-Aldrich), and β-tubulin (1:4,000 clone 2-28-33; T5293, Sigma-Aldrich). All secondary antibodies were HRP conjugated (Santa Cruz Biotechnology Inc.) and diluted 1:5,000 for rabbit-HRP, 1:3,000 for mouse-HRP, and 1:2,000 for goat-HRP.
Transfections. For transient expression, 293T cells were plated out at 2 × 105 cells per well on 6-well plates and transfected by 5 μl of TransFectin (Bio-Rad) complexed with 2 μg of MSCV-IRES-EGFP or MSCV-FLT3ITD-IRES-EGFP (mutant N51; ref. 24) and 0.5 μg of pcDNA3-WT C/EBPα (21). Five hours later, FLT3 inhibitors (0.5 μM MLN518 and 0.2 μM PKC-412) were added and the cells were cultured for an additional 16 hours. PD98059 (at 100 μM) was added 1.5 hours before the cell harvest, and TPA (at 10 nM) was added for the last 15 minutes of the culture. At the end of the treatments, cells were collected and lysed in 600 μl of 1× Laemmli Sample Buffer. Then 30 μl per lane were loaded on PAGE/SDS gels (7.5%).
Retroviral transductions. Retroviruses were produced by transfecting Phoenix A cells. Virus-containing supernatants were collected at 48 and 72 hours after transfection, filtered through 0.45-μm filter, and concentrated using a Centricon Plus-70 100000 MWCO column (Millipore).
Retroviral transduction was performed in culture dishes (Falcon 1008; BD) coated with 12 μg/ml RetroNectin during 2 consecutive days using a MOI between 2.5 and 5. The EGFP-expressing cells were enriched by sorting on day 3 and cultured for up to 8 additional days.
Lentiviral transductions. 293T cells were cotransfected using Lipofectamine 2000 with C/EBPα shRNA in pGhU6 vector or the shRNA control and lentiviral constructs Gag-Pol and Env. Virus was harvested and concentrated using a Centricon Plus-70 100000 MWCO column (Millipore). A single lentiviral transduction was performed in the presence of polybrene (8 μg/ml) (Sigma-Aldrich). MOLM-14 cells were infected with an MOI of 5. One day after transduction, cells were treated with either 0.01% DMSO control or 5 μM NU6102. Infected cells were determined by EGFP flow cytometry analysis.
Morphological examination. About 104 cells were spun at 500 g for 5 minutes onto glass slides and Wright-Giemsa stained with Diff-Quik solutions (Dade Behring).
Flow cytometry. Cells were washed once in PBS and blocked in 2% FBS/PBS on ice for 15 minutes. Surface staining was performed on ice for 30–40 minutes followed by 2 washes with PBS. Antibodies used were as follows: PE-conjugated anti-human CD11b (#555388; BD Biosciences — Pharmingen), PE-Cy5–conjugated anti-human CD11b (used in MOLM-14 cells transduced with pGhU6; #301308, Biolegend), FITC-conjugated anti-human CD11c (#11-0116-73; eBioscience), FITC-conjugated anti-human G-CSF–R/CD114 (# FAB381F; R&D Systems), APC-conjugated anti-human CD14 (# 561383; BD Biosciences), Pacific Blue–conjugated anti-human CD15 (#57-0159-73; eBioscience), eFluor605NC-conjugated anti-human CD16 (#93-0168-41, eBioscience), PE Cy7–conjugated anti-human CD33 (#25-0338-42, eBioscience), APC-conjugated anti-human CD34 (#343510, Biolegend), PE-Cy7–conjugated anti-human CD38 (#25-0389-41, eBioscience), and biotin-conjugated anti-human CD133 (#13-1338, eBioscience). Exclusion of dead cells was done by addition of DAPI. Cell sorting was performed using a FACSAria cell sorter, and immunophenotyping was done on an LSRII flow cytometer (BD Biosciences). Data were analyzed with FlowJo software (Treestar Inc.).
Quantitative RT-PCR. RNA was isolated by TRI Reagent (MRC Inc.), treated with DNaseI, and reverse-transcribed into cDNA (Invitrogen). Quantitative RT-PCR was performed using iQ Sybr Green Supermix (Bio-Rad). Amplification was done with a Corbett Rotor Gene 6000 (QIAGEN) using the following parameters: 95°C (10 minutes), 45 cycles of 95°C (15 s) and 60°C (1 minute). Primer sequences were as follows: human GAPDH F: 5′-CCACATCGCTCAGACACCAT-3′; human GAPDH R: 5′-CCAGGCGCCCAATACG-3′; human G-CSF-R F: 5′-TTTCAGGAACTTCTCTTGACGAGAA-3′; human G-CSF-R R: 5′-CGAGCCGAGCCTCAGTTTC-3′; human C/EBPε F: 5′-CTCCGATCTCTTTGCCGTGAA-3′; human C/EBPε R: 5′-TGGGCCGAAGGTATGTGGA-3′; human gelatinase A F: 5′-GTGGGACAAGAACCAGATCACAT-3′; human gelatinase A R: 5′-GTCTGCCTCTCCATCATGGATT-3′; human neutrophil elastase F: 5′-CCACCCGGCAGGTGTTC-3′; human neutrophil elastase R: 5′-GTGGCCGACCCGTTGAG-3′; human MPO F: 5′-AGACCTGCTGGAGAGGAA-3′; human MPO R: 5′-CGCAGCCGCTTGACTTG-3′; human lysozyme F: 5′-GCTGCAAGATAACATCGCT-3′; human lysozyme R: 5′-CCCATGCTCTAATGCCTTG-3′
Apoptosis assay. The analysis of apoptotic cells was performed using annexin V–FLUOS kit (Roche) according to the manufacturer’s protocol. Simultaneous labeling with propidium iodide (PI) was used for exclusion of the necrotic cells.
Cell-cycle analysis. Cells were suspended in phosphate-citrate buffer solution with 0.02% saponin for permeabilization and then incubated with 20 μg/ml Hoechst 33342 (Invitrogen) and 1 μg/ml Pyronin Y (Sigma-Aldrich). Incorporation of Hoechst 33342 and Pyronin Y were measured by flow cytometry.
Study approval. Patients’ informed consent was obtained in accordance with the Declaration of Helsinki. The study was approved by the Institutional Review Board: Committee on Clinical Investigations of Beth Israel Deaconess Medical Center.
-
Acknowledgments
We thank Karen O’Brien for useful suggestions and Christopher Hetherington for technical assistance. We also thank Yoshinobu Matsuo for MOLM-13, Neill Giese for MOLM-14, Stefan Heinrichs for MV4;11, and David Sternberg for Ba/F3-flt3ITD cell lines. Liu Yang provided CDK1 siRNA lentiviral constructs. We thank members of the Tenen and Gary Gilliland laboratories for many useful discussions and Mary Singleton and Toya Dessesaure for help in preparation of the manuscript. This research was supported by grants to D.G. Tenen from the NIH (P01 CA66996, P01 DK080665, and R01 CA 118316).
Address correspondence to: Daniel G. Tenen, Center for Life Sciences, 3 Blackfan Circle, Room 437, Boston, Massachusetts 02115, USA. Phone: 617.735.2205; Fax: 617.735.2222; E-mail: dtenen@bidmc.harvard.edu.
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2012;122(8):2955–2966. doi:10.1172/JCI43354.
Britta Will’s present address is: Albert Einstein College of Medicine, New York, New York, USA.
-
References
- Mrozek K, Heinonen K, Bloomfield CD. Clinical importance of cytogenetics in acute myeloid leukaemia. Best Pract Res Clin Haematol. 2001;14(1):19–47.
- Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3(2):89–101.
- Breitenbuecher F, et al. Identification of a novel type of ITD mutations located in nonjuxtamembrane domains of the FLT3 tyrosine kinase receptor. Blood. 2009;113(17):4074–4077.
- Yamamoto Y, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood. 2001;97(8):2434–2439.
- Chen P, Levis M, Brown P, Kim KT, Allebach J, Small D. FLT3/ITD mutation signaling includes suppression of SHP-1. J Biol Chem. 2005;280(7):5361–5369.
- Hayakawa F, et al. Tandem-duplicated Flt3 constitutively activates STAT5 and MAP kinase and introduces autonomous cell growth in IL-3-dependent cell lines. Oncogene. 2000;19(5):624–631.
- Kim KT, et al. Pim-1 is up-regulated by constitutively activated FLT3 and plays a role in FLT3-mediated cell survival. Blood. 2005;105(4):1759–1767.
- Mizuki M, et al. Flt3 mutations from patients with acute myeloid leukemia induce transformation of 32D cells mediated by the Ras and STAT5 pathways. Blood. 2000;96(12):3907–3914.View this article via: PubMed Google Scholar
- Mizuki M, et al. Suppression of myeloid transcription factors and induction of STAT response genes by AML-specific Flt3 mutations. Blood. 2003;101(8):3164–3173.
- Spiekermann K, Bagrintseva K, Schwab R, Schmieja K, Hiddemann W. Overexpression and constitutive activation of FLT3 induces STAT5 activation in primary acute myeloid leukemia blast cells. Clin Cancer Res. 2003;9(6):2140–2150.View this article via: PubMed Google Scholar
- Tse KF, Mukherjee G, Small D. Constitutive activation of FLT3 stimulates multiple intracellular signal transducers and results in transformation. Leukemia. 2000;14(10):1766–1776.
- Brown P, Small D. FLT3 inhibitors: a paradigm for the development of targeted therapeutics for paediatric cancer. Eur J Cancer. 2004;40(5):707–721.
- Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100(5):1532–1542.
- Nakao M, et al. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996;10(12):1911–1918.View this article via: PubMed Google Scholar
- Fathi A, Levis M. FLT3 inhibitors: a story of the old and the new. Curr Opin Hematol. 2011;18(2):71–76.
- Pratz KW, Sato T, Murphy KM, Stine A, Rajkhowa T, Levis M. FLT3-mutant allelic burden and clinical status are predictive of response to FLT3 inhibitors in AML. Blood. 2010;115(7):1425–1432.
- Ross SE, et al. Phosphorylation of C/EBPalpha inhibits granulopoiesis. Mol Cell Biol. 2004;24(2):675–686.
- Radomska HS, Huettner CS, Zhang P, Cheng T, Scadden DT, Tenen DG. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol Cell Biol. 1998;18(7):4301–4314.View this article via: PubMed Google Scholar
- Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci U S A. 1997;94(2):569–574.
- Qiao L, MacDougald OA, Shao J. CCAAT/enhancer-binding protein alpha mediates induction of hepatic phosphoenolpyruvate carboxykinase by p38 mitogen-activated protein kinase. J Biol Chem. 2006;281(34):24390–24397.
- Radomska HS, et al. Block of C/EBP alpha function by phosphorylation in acute myeloid leukemia with FLT3 activating mutations. J Exp Med. 2006;203(2):371–381.
- O’Farrell AM, et al. An innovative phase I clinical study demonstrates inhibition of FLT3 phosphorylation by SU11248 in acute myeloid leukemia patients. Clin Cancer Res. 2003;9(15):5465–5476.View this article via: PubMed Google Scholar
- Rocnik JL, et al. Roles of tyrosine 589 and 591 in STAT5 activation and transformation mediated by FLT3-ITD. Blood. 2006;108(4):1339–1345.
- Kelly LM, Liu Q, Kutok JL, Williams IR, Boulton CL, Gilliland DG. FLT3 internal tandem duplication mutations associated with human acute myeloid leukemias induce myeloproliferative disease in a murine bone marrow transplant model. Blood. 2002;99(1):310–318.
- Tamemoto H, et al. Biphasic activation of two mitogen-activated protein kinases during the cell cycle in mammalian cells. J Biol Chem. 1992;267(28):20293–20297.View this article via: PubMed Google Scholar
- Milella M, et al. Therapeutic targeting of the MEK/MAPK signal transduction module in acute myeloid leukemia. J Clin Invest. 2001;108(6):851–859.
- Ricciardi MR, et al. Quantitative single cell determination of ERK phosphorylation and regulation in relapsed and refractory primary acute myeloid leukemia. Leukemia. 2005;19(9):1543–1549.
- Desai D, Wessling HC, Fisher RP, Morgan DO. Effects of phosphorylation by CAK on cyclin binding by CDC2 and CDK2. Mol Cell Biol. 1995;15(1):345–350.View this article via: PubMed Google Scholar
- Pines J, Hunter T. Cyclins A and B1 in the human cell cycle. Ciba Found Symp. 1992;170:187–196.View this article via: PubMed Google Scholar
- Grundler R, Thiede C, Miething C, Steudel C, Peschel C, Duyster J. Sensitivity toward tyrosine kinase inhibitors varies between different activating mutations of the FLT3 receptor. Blood. 2003;102(2):646–651.
- Fiedler W, et al. A phase 2 clinical study of SU5416 in patients with refractory acute myeloid leukemia. Blood. 2003;102(8):2763–2767.
- Fiedler W, et al. A phase 1 study of SU11248 in the treatment of patients with refractory or resistant acute myeloid leukemia (AML) or not amenable to conventional therapy for the disease. Blood. 2005;105(3):986–993.
- Knapper S, et al. A phase 2 trial of the FLT3 inhibitor lestaurtinib (CEP701) as first-line treatment for older patients with acute myeloid leukemia not considered fit for intensive chemotherapy. Blood. 2006;108(10):3262–3270.
- Smith BD, et al. Single-agent CEP-701, a novel FLT3 inhibitor, shows biologic and clinical activity in patients with relapsed or refractory acute myeloid leukemia. Blood. 2004;103(10):3669–3676.
- Stone RM, et al. PKC 412 FLT3 inhibitor therapy in AML: results of a phase II trial. Ann Hematol. 2004;83(suppl 1):S89–S90.View this article via: PubMed Google Scholar
- Stone RM, et al. Patients with acute myeloid leukemia and an activating mutation in FLT3 respond to a small-molecule FLT3 tyrosine kinase inhibitor, PKC412. Blood. 2005;105(1):54–60.
- Piloto O, Wright M, Brown P, Kim KT, Levis M, Small D. Prolonged exposure to FLT3 inhibitors leads to resistance via activation of parallel signaling pathways. Blood. 2007;109(4):1643–1652.
- Heidel F, et al. Clinical resistance to the kinase inhibitor PKC412 in acute myeloid leukemia by mutation of Asn-676 in the FLT3 tyrosine kinase domain. Blood. 2006;107(1):293–300.
- Moore AS, et al. Selective FLT3 inhibition of FLT3-ITD(+) acute myeloid leukaemia resulting in secondary D835Y mutation: a model for emerging clinical resistance patterns [published online ahead of print February 22, 2012]. Leukemia. doi:10.1038/leu.2012.52. View this article via: PubMed Google Scholar
- DeAngelo DJ, et al. Phase 1 clinical results with tandutinib (MLN518), a novel FLT3 antagonist, in patients with acute myelogenous leukemia or high-risk myelodysplastic syndrome: safety, pharmacokinetics, and pharmacodynamics. Blood. 2006;108(12):3674–3681.
- Odgerel T, et al. The FLT3 inhibitor PKC412 exerts differential cell cycle effects on leukemic cells depending on the presence of FLT3 mutations. Oncogene. 2008;27(22):3102–3110.
- Behre G, et al. Ras signaling enhances the activity of C/EBP alpha to induce granulocytic differentiation by phosphorylation of serine 248. J Biol Chem. 2002;277(29):26293–26299.
- Ross SE, et al. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289(5481):950–953.
- Shim M, Smart RC. Lithium stabilizes the CCAAT/enhancer-binding protein alpha (C/EBPalpha) through a glycogen synthase kinase 3 (GSK3)-independent pathway involving direct inhibition of proteasomal activity. J Biol Chem. 2003;278(22):19674–19681.
- Geest CR, et al. p38 MAP kinase inhibits neutrophil development through phosphorylation of C/EBPalpha on serine 21. Stem Cells. 2009;27(9):2271–2282.
- Cha HC, et al. Phosphorylation of CCAAT/enhancer-binding protein alpha regulates GLUT4 expression and glucose transport in adipocytes. J Biol Chem. 2008;283(26):18002–18011.
- Satyanarayana A, Kaldis P. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene. 2009;28(33):2925–2939.
- Qian Z, Fernald AA, Godley LA, Larson RA, Le Beau MM. Expression profiling of CD34+ hematopoietic stem/ progenitor cells reveals distinct subtypes of therapy-related acute myeloid leukemia. Proc Natl Acad Sci U S A. 2002;99(23):14925–14930.
- Pan J, et al. TLS-ERG leukemia fusion protein deregulates cyclin-dependent kinase 1 and blocks terminal differentiation of myeloid progenitor cells. Mol Cancer Res. 2008;6(5):862–872.
- Matsuo Y, et al. Two acute monocytic leukemia (AML-M5a) cell lines (MOLM-13 and MOLM-14) with interclonal phenotypic heterogeneity showing MLL-AF9 fusion resulting from an occult chromosome insertion, ins(11;9)(q23;p22p23). Leukemia. 1997;11(9):1469–1477.
- Palacios R, Steinmetz M. Il-3-dependent mouse clones that express B-220 surface antigen, contain Ig genes in germ-line configuration, and generate B lymphocytes in vivo. Cell. 1985;41(3):727–734.
- van den Heuvel S, Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993;262(5142):2050–2054.
- Hirai H, et al. C/EBPbeta is required for ‘emergency’ granulopoiesis. Nat Immunol. 2006;7(7):732–739.
-
Version history
- Version 1 (July 17, 2012): No description
- Version 2 (August 1, 2012): No description



Copyright © 2025 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)










