Advertisement
Research ArticleCardiologyCell biology Free access | 10.1172/JCI122035
An alternative mitophagy pathway mediated by Rab9 protects the heart against ischemia
Toshiro Saito,1 Jihoon Nah,1 Shin-ichi Oka,1 Risa Mukai,1 Yoshiya Monden,1 Yasuhiro Maejima,2 Yoshiyuki Ikeda,1 Sebastiano Sciarretta,1 Tong Liu,3 Hong Li,3 Erdene Baljinnyam,1 Diego Fraidenraich,1 Luke Fritzky,4 Peiyong Zhai,1 Shizuko Ichinose,5 Mitsuaki Isobe,2 Chiao-Po Hsu,6 Mondira Kundu,7 and Junichi Sadoshima1
1Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
2Department of Cardiovascular Medicine, Tokyo Medical and Dental University, Tokyo, Japan.
3Center for Advanced Proteomics Research, Department of Biochemistry and Molecular Biology, and
4Core Imaging Facility, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
5Research Center for Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, Japan.
6Division of Cardiovascular Surgery, Department of Surgery, Taipei Veterans General Hospital, National Yang-Ming University School of Medicine, Taiwan.
7Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
Find articles by Saito, T. in: JCI | PubMed | Google Scholar
1Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
2Department of Cardiovascular Medicine, Tokyo Medical and Dental University, Tokyo, Japan.
3Center for Advanced Proteomics Research, Department of Biochemistry and Molecular Biology, and
4Core Imaging Facility, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
5Research Center for Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, Japan.
6Division of Cardiovascular Surgery, Department of Surgery, Taipei Veterans General Hospital, National Yang-Ming University School of Medicine, Taiwan.
7Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
Find articles by
Nah, J.
in:
JCI
|
PubMed
|
Google Scholar
|

1Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
2Department of Cardiovascular Medicine, Tokyo Medical and Dental University, Tokyo, Japan.
3Center for Advanced Proteomics Research, Department of Biochemistry and Molecular Biology, and
4Core Imaging Facility, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
5Research Center for Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, Japan.
6Division of Cardiovascular Surgery, Department of Surgery, Taipei Veterans General Hospital, National Yang-Ming University School of Medicine, Taiwan.
7Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
Find articles by
Oka, S.
in:
JCI
|
PubMed
|
Google Scholar
|

1Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
2Department of Cardiovascular Medicine, Tokyo Medical and Dental University, Tokyo, Japan.
3Center for Advanced Proteomics Research, Department of Biochemistry and Molecular Biology, and
4Core Imaging Facility, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
5Research Center for Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, Japan.
6Division of Cardiovascular Surgery, Department of Surgery, Taipei Veterans General Hospital, National Yang-Ming University School of Medicine, Taiwan.
7Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
Find articles by
Mukai, R.
in:
JCI
|
PubMed
|
Google Scholar
|

1Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
2Department of Cardiovascular Medicine, Tokyo Medical and Dental University, Tokyo, Japan.
3Center for Advanced Proteomics Research, Department of Biochemistry and Molecular Biology, and
4Core Imaging Facility, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
5Research Center for Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, Japan.
6Division of Cardiovascular Surgery, Department of Surgery, Taipei Veterans General Hospital, National Yang-Ming University School of Medicine, Taiwan.
7Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
Find articles by Monden, Y. in: JCI | PubMed | Google Scholar
1Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
2Department of Cardiovascular Medicine, Tokyo Medical and Dental University, Tokyo, Japan.
3Center for Advanced Proteomics Research, Department of Biochemistry and Molecular Biology, and
4Core Imaging Facility, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
5Research Center for Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, Japan.
6Division of Cardiovascular Surgery, Department of Surgery, Taipei Veterans General Hospital, National Yang-Ming University School of Medicine, Taiwan.
7Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
Find articles by Maejima, Y. in: JCI | PubMed | Google Scholar
1Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
2Department of Cardiovascular Medicine, Tokyo Medical and Dental University, Tokyo, Japan.
3Center for Advanced Proteomics Research, Department of Biochemistry and Molecular Biology, and
4Core Imaging Facility, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
5Research Center for Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, Japan.
6Division of Cardiovascular Surgery, Department of Surgery, Taipei Veterans General Hospital, National Yang-Ming University School of Medicine, Taiwan.
7Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
Find articles by Ikeda, Y. in: JCI | PubMed | Google Scholar
1Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
2Department of Cardiovascular Medicine, Tokyo Medical and Dental University, Tokyo, Japan.
3Center for Advanced Proteomics Research, Department of Biochemistry and Molecular Biology, and
4Core Imaging Facility, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
5Research Center for Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, Japan.
6Division of Cardiovascular Surgery, Department of Surgery, Taipei Veterans General Hospital, National Yang-Ming University School of Medicine, Taiwan.
7Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
Find articles by Sciarretta, S. in: JCI | PubMed | Google Scholar
1Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
2Department of Cardiovascular Medicine, Tokyo Medical and Dental University, Tokyo, Japan.
3Center for Advanced Proteomics Research, Department of Biochemistry and Molecular Biology, and
4Core Imaging Facility, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
5Research Center for Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, Japan.
6Division of Cardiovascular Surgery, Department of Surgery, Taipei Veterans General Hospital, National Yang-Ming University School of Medicine, Taiwan.
7Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
Find articles by Liu, T. in: JCI | PubMed | Google Scholar
1Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
2Department of Cardiovascular Medicine, Tokyo Medical and Dental University, Tokyo, Japan.
3Center for Advanced Proteomics Research, Department of Biochemistry and Molecular Biology, and
4Core Imaging Facility, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
5Research Center for Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, Japan.
6Division of Cardiovascular Surgery, Department of Surgery, Taipei Veterans General Hospital, National Yang-Ming University School of Medicine, Taiwan.
7Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
Find articles by Li, H. in: JCI | PubMed | Google Scholar
1Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
2Department of Cardiovascular Medicine, Tokyo Medical and Dental University, Tokyo, Japan.
3Center for Advanced Proteomics Research, Department of Biochemistry and Molecular Biology, and
4Core Imaging Facility, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
5Research Center for Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, Japan.
6Division of Cardiovascular Surgery, Department of Surgery, Taipei Veterans General Hospital, National Yang-Ming University School of Medicine, Taiwan.
7Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
Find articles by Baljinnyam, E. in: JCI | PubMed | Google Scholar
1Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
2Department of Cardiovascular Medicine, Tokyo Medical and Dental University, Tokyo, Japan.
3Center for Advanced Proteomics Research, Department of Biochemistry and Molecular Biology, and
4Core Imaging Facility, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
5Research Center for Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, Japan.
6Division of Cardiovascular Surgery, Department of Surgery, Taipei Veterans General Hospital, National Yang-Ming University School of Medicine, Taiwan.
7Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
Find articles by Fraidenraich, D. in: JCI | PubMed | Google Scholar
1Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
2Department of Cardiovascular Medicine, Tokyo Medical and Dental University, Tokyo, Japan.
3Center for Advanced Proteomics Research, Department of Biochemistry and Molecular Biology, and
4Core Imaging Facility, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
5Research Center for Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, Japan.
6Division of Cardiovascular Surgery, Department of Surgery, Taipei Veterans General Hospital, National Yang-Ming University School of Medicine, Taiwan.
7Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
Find articles by Fritzky, L. in: JCI | PubMed | Google Scholar
1Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
2Department of Cardiovascular Medicine, Tokyo Medical and Dental University, Tokyo, Japan.
3Center for Advanced Proteomics Research, Department of Biochemistry and Molecular Biology, and
4Core Imaging Facility, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
5Research Center for Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, Japan.
6Division of Cardiovascular Surgery, Department of Surgery, Taipei Veterans General Hospital, National Yang-Ming University School of Medicine, Taiwan.
7Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
Find articles by Zhai, P. in: JCI | PubMed | Google Scholar
1Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
2Department of Cardiovascular Medicine, Tokyo Medical and Dental University, Tokyo, Japan.
3Center for Advanced Proteomics Research, Department of Biochemistry and Molecular Biology, and
4Core Imaging Facility, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
5Research Center for Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, Japan.
6Division of Cardiovascular Surgery, Department of Surgery, Taipei Veterans General Hospital, National Yang-Ming University School of Medicine, Taiwan.
7Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
Find articles by Ichinose, S. in: JCI | PubMed | Google Scholar
1Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
2Department of Cardiovascular Medicine, Tokyo Medical and Dental University, Tokyo, Japan.
3Center for Advanced Proteomics Research, Department of Biochemistry and Molecular Biology, and
4Core Imaging Facility, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
5Research Center for Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, Japan.
6Division of Cardiovascular Surgery, Department of Surgery, Taipei Veterans General Hospital, National Yang-Ming University School of Medicine, Taiwan.
7Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
Find articles by Isobe, M. in: JCI | PubMed | Google Scholar
1Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
2Department of Cardiovascular Medicine, Tokyo Medical and Dental University, Tokyo, Japan.
3Center for Advanced Proteomics Research, Department of Biochemistry and Molecular Biology, and
4Core Imaging Facility, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
5Research Center for Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, Japan.
6Division of Cardiovascular Surgery, Department of Surgery, Taipei Veterans General Hospital, National Yang-Ming University School of Medicine, Taiwan.
7Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
Find articles by Hsu, C. in: JCI | PubMed | Google Scholar
1Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
2Department of Cardiovascular Medicine, Tokyo Medical and Dental University, Tokyo, Japan.
3Center for Advanced Proteomics Research, Department of Biochemistry and Molecular Biology, and
4Core Imaging Facility, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
5Research Center for Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, Japan.
6Division of Cardiovascular Surgery, Department of Surgery, Taipei Veterans General Hospital, National Yang-Ming University School of Medicine, Taiwan.
7Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
Find articles by Kundu, M. in: JCI | PubMed | Google Scholar
1Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
2Department of Cardiovascular Medicine, Tokyo Medical and Dental University, Tokyo, Japan.
3Center for Advanced Proteomics Research, Department of Biochemistry and Molecular Biology, and
4Core Imaging Facility, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
5Research Center for Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo, Japan.
6Division of Cardiovascular Surgery, Department of Surgery, Taipei Veterans General Hospital, National Yang-Ming University School of Medicine, Taiwan.
7Department of Pathology, St. Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
Find articles by Sadoshima, J. in: JCI | PubMed | Google Scholar
Published December 4, 2018 - More info
J Clin Invest. 2019;129(2):802–819. https://doi.org/10.1172/JCI122035.
Copyright © 2019, American Society for Clinical Investigation
Received: May 10, 2018; Accepted: November 27, 2018
Related article:
Abstract
The heart relies on mitochondria-derived energy production for continuous contraction and relaxation; therefore, the maintenance of a pool of healthy mitochondria is essential for sustaining normal cardiac performance. Mitophagy serves as a critical process for maintaining mitochondrial quality control and involves the PTEN-induced kinase 1/Parkin (Pink1/Parkin) pathway and autophagosomes labeled with the autophagy proteins autophagy-related 7 (ATG) and light chain 3 (LC3). In this issue of the JCI, Saito and colleagues identify an alternative pathway for mitophagy that utilizes the serine/threonine protein kinase Unc-51–like kinase 1 (Ulk1) and the small GTPase Rab9 to clear damaged mitochondria independently of conventional autophagy proteins. Together, the results of this study reveal that Ulk1 phosphorylation of Rab9 at serine 179 is critical for alternative mitophagy and cardioprotection under energy stress conditions.
Authors
Rimpy Dhingra, Inna Rabinovich-Nikitin, Lorrie A. Kirshenbaum
-
Abstract
Energy stress, such as ischemia, induces mitochondrial damage and death in the heart. Degradation of damaged mitochondria by mitophagy is essential for the maintenance of healthy mitochondria and survival. Here, we show that mitophagy during myocardial ischemia was mediated predominantly through autophagy characterized by Rab9-associated autophagosomes, rather than the well-characterized form of autophagy that is dependent on the autophagy-related 7 (Atg) conjugation system and LC3. This form of mitophagy played an essential role in protecting the heart against ischemia and was mediated by a protein complex consisting of unc-51 like kinase 1 (Ulk1), Rab9, receptor-interacting serine/thronine protein kinase 1 (Rip1), and dynamin-related protein 1 (Drp1). This complex allowed the recruitment of trans-Golgi membranes associated with Rab9 to damaged mitochondria through S179 phosphorylation of Rab9 by Ulk1 and S616 phosphorylation of Drp1 by Rip1. Knockin of Rab9 (S179A) abolished mitophagy and exacerbated the injury in response to myocardial ischemia, without affecting conventional autophagy. Mitophagy mediated through the Ulk1/Rab9/Rip1/Drp1 pathway protected the heart against ischemia by maintaining healthy mitochondria.
-
Introduction
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved mechanism that degrades cytoplasmic macromolecules, including misfolded proteins and damaged organelles, to preserve cellular homeostasis (1). Increasing lines of evidence suggest that autophagy can degrade specific proteins and organelles in a context-dependent manner. For example, unnecessary, depolarized, or disorganized mitochondria are eliminated by a mitochondria-specific form of autophagy, termed mitophagy, to maintain the quality of this organelle, which is essential for ATP production but, upon damage, can be a source of ROS and trigger programmed cell death (2). The molecular mechanisms by which autophagy degrades specific targets have been the subject of intense research during the past decade, and several important mechanisms have been elucidated in mammalian cells (3).
One of the most well-defined mechanisms through which damaged mitochondria are degraded by autophagy (hereafter called mitophagy) is the Pink1-Parkin–dependent mechanism (4). When mitochondria are damaged and depolarized, Pink1, a mitochondrial kinase, is stabilized and recruits Parkin, an E3 ligase, to mitochondria through phosphorylation of either ubiquitin (5, 6) or mitofusin 2 (Mfn2) (7). Pink1 alone or K63-linked ubiquitin chains formed on the outer mitochondrial membrane recruit LC3 receptors or adapters, including NDP52, optineurin, p62 (also known as SQSTM1), NBR1, and NIX, thereby promoting autophagosome formation, a process facilitated by phosphorylation of ubiquitin and Parkin by Pink1 (8). These mechanisms allow damaged mitochondria to be engulfed by autophagosomes through LC3-dependent mechanisms. Although these mechanisms mediate mitophagic degradation of mitochondria under some conditions in vitro, including depolarization by mitochondrial uncouplers, whether these mechanisms also mediate mitophagy in vivo in response to more pathophysiologically relevant stimuli, such as ischemia, remains to be shown. Importantly, increasing evidence suggests that mitochondria can be degraded by multiple pathways, including Parkin-independent mechanisms of autophagy (9) and autophagy-related 5– (Atg5-) and Atg7-independent, nonconventional autophagy (10, 11).
The heart is a highly energy-demanding organ because of its sustained contraction and relaxation, and thus mitochondria are abundant in cardiomyocytes (CMs). Mitochondria produce the majority of ATP in CMs, together with physiological production of ROS. However, under pathological conditions, excessive ROS damage mitochondrial proteins, exacerbating ROS production and activating the cell death pathway. Accordingly, efficient clearance of dysfunctional mitochondria during stress in CMs is essential to preserve their viability and functionality. Interestingly, mitochondria in adult CMs have several unique features. First, they are fragmented compared with mitochondria in other cell types, rather than interconnected, and compacted (12). Second, the turnover of cardiac mitochondrial proteins is slower than that of cytosolic proteins (13). Thus, it is possible that CMs may possess unique mechanisms to maintain the quality of mitochondria.
We have shown recently that mitophagy plays an essential role in preventing mitochondrial dysfunction and cardiac dysfunction during pressure overload in the heart in vivo (14). Interestingly, mitophagy is activated after the conventional form of autophagy is inactivated, suggesting that mitophagy in the heart may be mediated by a mechanism distinct from the conventional mechanisms of autophagy (14). In fact, we subsequently discovered that an Atg7-independent form of autophagy plays a predominant role in mediating mitochondrial degradation in the heart in response to starvation.
The present study thus aimed to clarify the molecular mechanism that mediates mitophagy during pathophysiological stress in the heart. We used both starvation and hypoxia as stimuli to induce mitophagy in the heart and the CMs therein. We asked: (a) What is the predominant signaling mechanism mediating mitophagy during energy stress in the heart? and (b) What is the molecular mechanism by which damaged mitochondria are recognized by autophagosomes during energy stress in the heart?
-
Results
Mitophagy is induced by energy stress via an Atg7-independent but Ulk1-dependent mechanism in the heart. In order to investigate the molecular mechanism of mitophagy in the heart, key components of the autophagy machinery were downregulated by Myh6-Cre recombinase–driven genetic ablation of the floxed gene of interest. Forty-eight hours of starvation served to impose energy stress and induce autophagy and mitophagy in the heart (15, 16). Conventional autophagy, evaluated by measuring LC3-II protein levels, was increased significantly in the hearts of WT and cardiac-specific Ulk1-knockout mice (Ulk1-cKO) during starvation (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI122035DS1). Autophagic flux, evaluated following chloroquine injection, was preserved in WT and Ulk1-cKO hearts during starvation (Figure 1, A and B). In contrast, in the hearts of cardiac-specific Atg7-knockout mice (Atg7-cKO), LC3-II expression was not induced, and a marked accumulation of p62 was observed at baseline and during starvation (Supplemental Figure 1A). LC3-II expression was not increased at baseline or after starvation, even in the presence of chloroquine, suggesting that autophagic flux was completely inhibited in the hearts of Atg7-cKO mice during starvation (Figure 1, A and B). Protein levels of Ulk1 in Atg7-cKO mice and of Parkin in Ulk1-cKO mice were increased at baseline and during starvation (Supplemental Figure 1A), which may indicate the activation of compensatory mechanisms. Despite the marked suppression of autophagy and autophagic flux in Atg7-cKO hearts, electron microscopic (EM) analyses showed mitochondria engulfed by double-membrane structures with mitophagic characteristics in the hearts of WT and Atg7-cKO mice during starvation (Supplemental Figure 1, B and C). Furthermore, when transgenic mice with cardiac-specific expression of Mito-Keima, a reliable indicator of mitophagy (17), were crossed with Atg7-cKO mice (Atg7-cKO/Mito-Keima–Tg) and subjected to starvation, we detected an increase in the area of dots with high 560 nm/440 nm ratios, indicating lysosomal localization of mitochondrial proteins, even in the absence of Atg7 (Figure 1, C and D), indicating that autophagosomes containing mitochondria in Atg7-cKO mice mature into autolysosomes. Interestingly, autophagosomes and autolysosomes containing mitochondria were rarely detected in Ulk1-cKO hearts (Supplemental Figure 1, B and C). The induction of dots with high ratios of Mito-Keima signal during starvation was significantly attenuated in Mito-Keima–Tg mice crossed with Ulk1-cKO mice (Ulk1-cKO/Mito-Keima–Tg) compared with that seen in control Mito-Keima–Tg mice (WT/Mito-Keima–Tg) (Figure 1, C and D). Consistent with these data, mitochondrial content, as evaluated by mitochondrial DNA (mtDNA) copy numbers, was significantly decreased during starvation in WT and Atg7-cKO hearts but was increased in Ulk1-cKO hearts, suggesting that mitochondrial degradation takes place in WT and Atg7-cKO hearts during starvation, whereas mitochondrial degradation may be impaired in Ulk1-cKO hearts (Supplemental Figure 1D).
 Figure 1
Figure 1Ulk1- but not Atg7-dependent autophagy protects the heart against physiological energy stress through activation of mitophagy. Mice were subjected to 48 hours of starvation. Error bars indicate the SEM. (A and B) Protein levels of LC3 in the heart were analyzed after chloroquine administration (10 μg/kg i.p.) during the 48-hour starvation period to evaluate autophagic flux. (A) Representative immunoblot and (B) summary of relative expression of LC3-II (n = 7–8 per group). #P < 0.05 versus WT, control, and no chloroquine; †P < 0.05 versus WT, starvation, and no chloroquine; *P < 0.05 versus Ulk1-cKO, control, and no chloroquine; ‡P < 0.05 versus Ulk1-cKO, starvation, and no chloroquine (paired Student’s t test). (C and D) Lysosomal degradation of mitochondria was examined in the hearts of Mito-Keima–Tg mice during starvation. The bright signal corresponding to a high 560/440-nm ratio shows lysosomal localization of Mito-Keima protein, indicating mitophagy. (C) Representative images are shown. Scale bars: 50 μm (top); original magnification, ×3 (enlarged insets, bottom). (D) Summary of signal area/cell area ratio (n = 3 per group). #P < 0.05 versus WT and control; †P < 0.01 versus WT and starvation; *P < 0.01 versus Atg7-cKO and control; §P < 0.05 versus Atg7-cKO and starvation (Tukey-Kramer’s test). CQ, chloroquine; Ctr, control; Stv, starvation.
We also investigated whether activation of mitophagy during energy stress is Parkin dependent in the heart. The hearts of Parkin systemic KO mice (Parkin-KO) crossed with Mito-Keima-Tg mice showed an increase in the area of dots with high 560 nm/440 nm ratios of Mito-Keima during starvation, indicating that starvation-induced mitophagy in the heart is Parkin independent (Supplemental Figure 1, E and F).
Taken together, these results suggest that mitophagy activated by starvation in the heart is distinct from the well-characterized Atg7- and Parkin-dependent form of mitophagy observed in many cell types and is instead characterized by Ulk1 dependency.
Ulk1-dependent, but not Atg7-dependent, mechanisms protect the heart against prolonged ischemia. In order to further elucidate the role of Ulk1-dependent mechanisms in protecting the heart during energy stress, mice were subjected to myocardial ischemia. LC3-dependent autophagy was induced in WT and Ulk1-cKO mice but not in Atg7-cKO mice over a 30-minute period of myocardial ischemia (Figure 2A). EM analyses showed that the abundance of mitochondria engulfed by autophagosomes was increased in WT and Atg7-cKO CMs during the 30-minute period of ischemia but that this engulfment was impaired in Ulk1-cKO CMs (Supplemental Figure 2, A and B). Analysis with Atg7-cKO/Mito-Keima–Tg mice displayed an increase in the area of dots with high 560 nm/440 nm ratios of Mito-Keima during the 30-minute period of ischemia, indicating lysosomal localization of mitochondrial proteins (Figure 2, B and C, and Supplemental Figure 2, C and D). The high-ratio dots were clustered in the perinuclear area in WT/Mito-Keima–Tg and Atg7-cKO/Mito-Keima–Tg hearts (Supplemental Figure 2D). Moreover, the content of autolysosomes appeared to undergo degradation during the 30 minutes of ischemia, even in the absence of Atg7, as indicated by the presence of partially degraded mitochondria within autolysosomes (Supplemental Figure 2E). However, there were fewer dots with high 560 nm/440 nm ratios in Ulk1-cKO/Mito-Keima–Tg hearts than in WT/Mito-Keima–Tg and Atg7-cKO/Mito-Keima–Tg hearts during the 30-minute period of ischemia (Figure 2, B and C, and Supplemental Figure 2, C and D). The mitochondrial content, as evaluated with COX I/GAPDH, was significantly decreased in WT and Atg7-cKO hearts but was increased in Ulk1-cKO hearts during the 30 minutes of ischemia, consistent with the impairment of mitochondrial degradation in Ulk1-cKO but not Atg7-cKO hearts (Supplemental Figure 2, F and G). After 2 hours of ischemia, the percentage of infarct size/area at risk (AAR) was significantly greater in Ulk1-cKO hearts than in WT hearts (Figure 2, D and E). These results suggest that Ulk1, rather than Atg7, mediates mitophagy during prolonged ischemia in the heart in vivo. Furthermore, these findings indicate that endogenous Ulk1, rather than Atg7, plays an essential role in protecting the heart against ischemic injury.
 Figure 2
Figure 2Ulk1- but not Atg7-dependent autophagy protects the heart against ischemia through activation of mitophagy. Mice were subjected to prolonged ischemia. Hearts were excised at the indicated time points after coronary artery ligation. Error bars represent the SEM. (A) Protein levels of LC3 in the heart were analyzed. (B and C) Lysosomal degradation of mitochondria was examined in the hearts of Mito-Keima–Tg mice during ischemia. Scale bars: 50 μm (top); original magnification, ×3 (enlarged insets, bottom). (B) Representative images are shown. (C) Summary of signal area/cell area percentage (n = 3–4 per group). #P < 0.01 versus WT; *P < 0.01 versus Atg7-cKO (Tukey-Kramer’s test). (D) Representative images of LV slices with Alcian blue and TTC staining. Scale bars: 5 mm. (E) AAR/LV and infarct size/AAR ratios. n = 5–6 per group. #P < 0.05 versus WT (Tukey-Kramer’s test).
Mitophagy is induced by energy stress via an Atg7-independent but Ulk1-dependent mechanism in CMs. To examine the molecular mechanism by which mitophagy is activated in response to energy stress in CMs, we knocked down Atg7 and Ulk1 in cultured CMs using adenoviruses harboring shRNA against Atg7 (Ad-sh-Atg7) and Ulk1 (Ad-sh-Ulk1), respectively, at baseline (Supplemental Figure 3A), and the cells were subjected to 4 hours of either glucose deprivation (GD) or hypoxia. We observed no compensatory upregulation of Ulk1 or Parkin in the CMs with sh-Atg7 or sh-Ulk1, respectively (Supplemental Figure 3A).
Consistent with the data obtained in vivo, we observed mitochondria enclosed by autophagosomes in EM analyses of CMs treated with sh-control or sh-Atg7 during GD (Supplemental Figure 3B). To quantify lysosomal degradation of mitochondria, we transduced CMs with Mito-Keima (16, 17). GD or hypoxia significantly increased the area of dots with high 560 nm/440 nm ratios, indicative of the presence of mitochondrial protein in lysosomes, in CMs treated with sh-control or sh-Atg7 (Figure 3, A and B, and Supplemental Figure 3C), suggesting that mitophagy can be observed during energy stress in CMs, even in the presence of marked downregulation of Atg7. Interestingly, the number of mitochondria enclosed by autophagosomes, as evaluated with EM analyses (Supplemental Figure 3B), or degraded by lysosomes, as evaluated with Mito-Keima (Figure 3, A and B, and Supplemental Figure 3C), in the presence of GD or hypoxia was significantly decreased in the presence of sh-Ulk1. The loss of mitochondrial membrane potential, as evaluated with 5,5′,6,6′- tetrachloro-1,1′,3,3′-tetraethylbenzimidazolocarbocyanine iodide (JC-1) and tetramethylrhodamine ethyl ester (TMRE) staining, was more prominent in CMs treated with Ad-sh-Ulk1 alone or with combined Ad-sh-Atg7 and Ad-sh-Ulk1 than in those treated with Ad-sh-control or Ad-sh-Atg7 during GD (Supplemental Figure 3D). Furthermore, we observed that cell death was greater in CMs treated with Ad-sh-Ulk1 than in those treated with Ad-sh-control or Ad-sh-Atg7 during GD (Supplemental Figure 3E). These results are consistent with the notion that Ulk1-dependent autophagy, but not Atg7-dependent autophagy, protects CMs against energy stress through activation of mitophagy. Since endogenous Parkin was upregulated in the hearts of Ulk1-cKO mice (Supplemental Figure 1A), it is possible that Parkin might contribute to the greater infarction seen in Ulk1-cKO mice during ischemia (Figure 2, D and E). In order to clarify the effect of Parkin in the absence of Ulk1, we analyzed the cellular viability of CMs treated with Ad-sh-control or Ad-sh-Ulk1 during hypoxia (Supplemental Figure 3F). In a result similar to what was observed in vivo, we found that transduction with Ad-sh-Ulk1 exacerbated hypoxia-induced CM death compared with control. Although downregulation of Ulk1 alone did not affect the level of Parkin in CMs in vitro, hypoxia-induced upregulation of Parkin was enhanced in the absence of Ulk1. We observed that normalization of Parkin levels with Ad-sh-Parkin did not significantly affect the Ad-sh-Ulk1–induced exacerbation of hypoxia-induced CM death (Supplemental Figure 3F), suggesting that the greater infarction in Ulk1-cKO hearts during ischemia was not mediated by upregulation of Parkin.
 Figure 3
Figure 3Atg7-independent and Ulk1-dependent autophagy is the predominant form of mitophagy induced by energy stress in CMs. Lysosomal degradation of mitochondria was examined in CMs transduced with Mito-Keima and subjected to 4 hours of GD or hypoxia (HO). Error bars represent the SD. (A) Representative images showing a high 560/440-nm ratio of dots. Scale bars: 50 μm (top); original magnification, ×3.8 (enlarged insets, bottom) . (B) Quantitation of the area with a high ratio of dots per cell area (percentage). Number of cells in each group: 134, 46, and 50 (sh-Control); 23, 19, and 28 (sh-Atg7); and 18, 25, and 31 (sh-Ulk1) from 3 independent experiments. #P < 0.01 versus sh-control and no treatment; †P < 0.01 versus sh-control and GD; *P < 0.01 versus sh-control and hypoxia; ‡P < 0.01 versus sh-Atg7 and no treatment (Tukey-Kramer’s test). NT, no treatment.
Energy stress induces Rab9-dependent mitophagy in CMs. We then explored the possibility that the Atg7-independent mitophagy described above is mediated through the alternative autophagy originally reported by Nishida et al. In alternative autophagy, autophagosomes are derived from trans-Golgi and endosomes and associate with Rab9 but not LC3 (18). We examined the effect of brefeldin A (BFA), which inhibits Golgi-derived membranes, upon mitophagy in CMs. BFA significantly reduced the area of high-ratio Mito-Keima dots during GD, but induction of conventional autophagy, as indicated by increases in LC3-II, was not affected (Supplemental Figure 3, G and H). On the other hand, we found that the high-ratio dots induced by overexpression of Parkin, which presumably induces Parkin-mediated mitophagy, were not significantly inhibited by BFA (Supplemental Figure 3, I–K). These results suggest that the mitophagy induced by energy stress in CMs has a unique feature, namely BFA sensitivity, and is distinct from Parkin-mediated mitophagy. They also suggest that the autophagosomes engulfing mitochondria observed in CMs during starvation originate from Golgi-derived membranes.
To further elucidate the role of Rab9 in mediating mitophagy, we transduced CMs with fluorescent protein–tagged Rab9 (yellow fluorescent protein–Rab9 [YFP-Rab9] or red fluorescent protein–Rab9 [RFP-Rab9]). We found that GD for 4 hours significantly increased the number of YFP-Rab9 puncta and that the GD-induced increases in YFP-Rab9 puncta were suppressed in CMs with either Ulk1 or Beclin1 downregulation. Inhibition of autophagosome-lysosome fusion by bafilomycin A1 further increased YFP-Rab9 puncta during GD, indicating that the puncta were degraded in a lysosome-dependent manner (Supplemental Figure 4, A and B). Consistently, endogenous Rab9 colocalized with Lamp2, a lysosomal marker, during GD in a Ulk1-dependent manner (Supplemental Figure 4C). A significant subpopulation of Lamp2-positive puncta may represent Lamp2A-positive lysosomes that remain active for chaperone-mediated autophagy (CMA). However, during starvation, colocalization of Rab9 puncta with Lamp2A puncta did not increase in CMs (Supplemental Figure 4D), suggesting that the changes in total Lamp2 staining and its association with Rab9 do not reflect CMA in CMs and reliably reflect macroautophagy. These results suggest that Rab9 associates with autophagosomes or autolysosomes during GD in a Ulk1- and Beclin1-dependent manner. In order to test whether Rab9 puncta contain mitochondria, we examined the colocalization of YFP-Rab9 with Mito-Keima. We observed that the ring structure of YFP-Rab9 enclosed Mito-Keima dots with high 560 nm/440 mm ratios during GD, indicating that YFP-Rab9 colocalizes with autolysosomes containing mitochondria (Figure 4A and Supplemental Figure 4E). Importantly, most of Mito-Keima dots with high ratios merged with YFP-Rab9 puncta, suggesting that mitophagy predominantly occurs through Rab9-positive autolysosomes during energy stress in CMs. In the presence of bafilomycin A1, however, the ring structure of YFP-Rab9 encircled neutral Mito-Keima dots during GD (Supplemental Figure 4, F and G). Line-scan analyses of the confocal images indicated that Mito-Keima and YFP-Rab9 were colocalized (Supplemental Figure 4G), suggesting that YFP-Rab9 colocalizes with autophagosomes containing mitochondria. Consistently, the ring structure of YFP-Rab9 (WT) also colocalized with DsRed-Mito, another tracker of mitochondria, during GD in the presence of chloroquine (Figure 4B, left). Under the same experimental conditions, however, GFP-LC3 did not colocalize with either DsRed-Mito or RFP-Rab9 (Supplemental Figure 4, H and I).
 Figure 4
Figure 4Evaluation of Rab9 puncta in CMs in vitro. (A) Representative confocal micrographs of CMs transduced with Mito-Keima and YFP-Rab9 during GD. Scale bars: 10 μm; original magnification, ×2.5 (enlarged insets, bottom). (B) Representative confocal micrographs of CMs transduced with YFP-Rab9 (WT or S179A) and DsRed-Mito during GD with chloroquine. Scale bars: 10 μm; original magnification, ×2.4 (enlarged insets, bottom).
In order to further confirm that Rab9 associates with the mitophagy observed in CMs, we conducted Immunogold EM analyses. Autophagosomes induced by GD in control CMs and in CMs in which Atg7 was downregulated, but not in CMs in which Ulk1 was downregulated, were labeled with Rab9 antibody (Supplemental Figure 5). Although autophagosomes induced by GD in control CMs and in CMs in which Ulk1 was downregulated were labeled with LC3 antibody, as expected, those in CMs in which Atg7 was downregulated were not labeled (Supplemental Figure 5).
In order to clarify the function of Rab9 in mediating mitophagy in the heart, we generated transgenic mice with cardiac-specific expression of YFP-Rab9 (YFP-Rab9–Tg mice) and obtained bigenic mice by crossing YFP-Rab9–Tg mice with Mito-Keima–Tg mice (Mito-Keima–Tg/YFP-Rab9–Tg). The hearts of WT/Mito-Keima–Tg/YFP-Rab9–Tg mice showed YFP-Rab9 puncta during the 30-minute period of ischemia (Figure 5, arrows). These YFP-Rab9 puncta appeared to cluster around the perinuclear region (Figure 5, arrowheads). As expected, mitophagy, indicated by Mito-Keima dots with high 560 nm/440 nm signal ratios, were primarily colocalized with YFP-Rab9 (Figure 5). Since Rab9 puncta are distinct from LC3-positive autophagosomes (Supplemental Figure 4I), these data suggest that the hearts of WT mice preferentially utilize Rab9-positive autophagosomes, even in the presence of LC3-positive autophagosomes, to mediate mitophagy during ischemia. Consistent with the data obtained in vitro, deletion of Ulk1 (Ulk1-cKO/Mito-Keima–Tg/YFP-Rab9–Tg) suppressed both Rab9 puncta and mitophagy signals in the heart (Figure 5).
 Figure 5
Figure 5Evaluation of Rab9 puncta in the heart. Colocalization of Rab9 puncta with mitophagy signal was examined in the hearts of Mito-Keima–Tg/YFP-Rab9–Tg mice during ischemia. Representative confocal micrographs are shown. Scale bars: 50 μm; original magnification, ×3 (enlarged insets). Arrows and arrowheads indicate puncta and clusters of YFP-Rab9, respectively.
Finally, in order to test the causative involvement of Rab9 in mitophagy induced by energy stress in CMs, Rab9 was downregulated with sh-Rab9. We found that sh-Rab9 significantly suppressed the increases in high-ratio Mito-Keima dots induced by GD or hypoxia (Figure 6, A and B, and Supplemental Figure 6A), indicating that Rab9 plays an essential role in mediating the mitophagy induced by energy stress in CMs. Downregulation of either Ulk1 or Rab9 significantly impaired ATP production in mitochondria isolated from CMs during GD, suggesting that endogenous Ulk1 and Rab9 are required to maintain mitochondrial function during energy stress (Supplemental Figure 6B).
 Figure 6
Figure 6Ulk1 induces mitophagy through Rab9 and Drp1 in response to energy stress in CMs. Lysosomal degradation of mitochondria was examined in CMs transduced with Mito-Keima. (A and C) Representative images showing high ratios of Mito-Keima dots. Scale bars: 50 μm; original magnification, ×2.8 (enlarged insets, bottom). (B) Quantitation of the area with a high ratio of dots per total cell area. Number of cells in each group: 69, 29, and 19 (sh-control) and 18, 25, and 40 (sh-Rab9) from 3 independent experiments. #P < 0.01 versus sh-control and no treatment; †P < 0.01 versus sh-control and GD; *P < 0.05 versus sh-control and hypoxia (Tukey-Kramer’s test). (D) Quantitation of the area with a high ratio of dots per total cell area. Number of cells in each group: 69 (LacZ/sh-control), 53 (Ulk1/sh-control), 15 (Ulk1/sh-Rab9), and 29 (Ulk1/sh-Drp1). #P < 0.01 versus LacZ and sh-control; †P < 0.01 versus Ulk1 and sh-control (Tukey-Kramer’s test). Error bars represent the SD.
Taken together, these observations indicate that Rab9, but not LC3, colocalizes with autophagosomes and autolysosomes containing mitochondria in a Ulk1-dependent manner during energy stress in CMs, suggesting that alternative autophagy is the predominant form of mitophagy in CMs during energy stress.
Mitophagy in response to energy stress is mediated through a Drp1-dependent mechanism in CMs. We have previously shown that Drp1, a mammalian homolog of Dnm1, plays a critical role in mediating mitophagy during energy stress in the heart (16). Dnm1, a mitochondrial fission protein, plays an essential role in mediating mitophagy during nutrient starvation in yeast (19). Thus, we hypothesized that Drp1 plays essential roles in mediating Ulk1-dependent mitophagy in CMs. The high ratio of Mito-Keima dots induced by overexpression of Ulk1 was suppressed by either sh-Rab9 or sh-Drp1 (Figure 6, C and D, and Supplemental Figure 6C), suggesting that Ulk1 induces mitophagy through both Rab9 and Drp1. Downregulation of Rab9 also suppressed GD-induced phosphorylation of Drp1 and decreases in the size of individual mitochondria in CMs (Supplemental Figure 6, D and E), suggesting that Rab9 may also affect mitochondrial fission and fusion.
Since Ulk1 is a serine/threonine kinase and mitochondrial translocation of Drp1 is induced by phosphorylation at S616 (20, 21), we investigated whether phosphorylation of Drp1 at S616 is mediated by endogenous Ulk1. We found that sh-Ulk1 suppressed S616 phosphorylation of Drp1 during GD, whereas overexpression of Ulk1 upregulated Drp1 S616 phosphorylation in CMs (Supplemental Figure 7, A and B). Similarly, although phosphorylation of Drp1 at S616 was significantly increased in WT and Atg7-cKO hearts in response to starvation, it was abolished in Ulk1-cKO hearts (Figure 7, A and B). Since mitochondrial translocation of Drp1 may mediate mitochondrial fission, we evaluated the size of individual mitochondria by EM analyses. The average cross-sectional area of a single mitochondrion was significantly decreased in WT and Atg7-cKO CMs during starvation, but was unaffected in Ulk1-cKO CMs (Supplemental Figure 7, C and D). These results are consistent with the notion that Ulk1 plays an essential role in mediating mitochondrial fission through S616 phosphorylation of Drp1 during energy stress. Interestingly, however, recombinant Ulk1 failed to phosphorylate Drp1 at S616 in the test tube (Supplemental Figure 7E). Thus, another kinase regulated by Ulk1 presumably phosphorylates Drp1.
 Figure 7
Figure 7Ulk1, Rab9, and Rip1, a Drp1 kinase, form a functional complex. (A and B) Mice were subjected to 48 hours of starvation. Phosphorylation of Drp1 at S616 in the heart was analyzed. Representative immunoblots are shown in A, and a summary is shown in B (n = 7–8 per group). Error bars represent the SEM. #P < 0.05 versus WT and control; *P < 0.05 versus Atg7-cKO and control (paired Student’s t test). (C) Co-IP was performed in CMs. Right: Ulk1 was detected on a parallel gel using the same samples. (D) The heart lysate was assayed using high-pressure chromatography and a size-exclusion column, as shown in Supplemental Figure 7. Co-IP was conducted in fractions 4–6 from the heart lysate of WT/YFP-Rab9–Tg or Ulk1-cKO/YFP-Rab9–Tg mice subjected to starvation.
Rip1 mediates the phosphorylation of Drp1 at S616 during energy stress. We then hypothesized that Ulk1, Rab9, and Drp1 form a functional complex, which in turn facilitates mitochondrial fission and autophagy during energy stress. Co-IP assays indicated that Ulk1 interacts with YFP-Rab9 in CMs (Figure 7C). Since Ulk1 does not directly phosphorylate Drp1, we investigated whether known Drp1 kinases form a protein complex with Ulk1 and Rab9. CDK1, CDK5, ERK2, and Rip1 have been reported to act as Drp1 kinases (20–23). Of these, Rip1 was found to be associated with YFP-Rab9 in CMs (Figure 7C and Supplemental Figure 7F). Rip1 directly phosphorylates Drp1 at S616 and interacts with Drp1 in bone marrow–derived macrophages (20). We found that Rip1 bound to YFP-Rab9 in CMs and that the association of Rip1 with YFP-Rab9 was Ulk1 dependent (Figure 7C). To examine the interaction of Rip1 with Drp1 in CMs, we performed co-IP assays. We found that Rip1 interacted with HA-Drp1 in a Ulk1- and Rab9-dependent manner (Supplemental Figure 7G). Likewise, in co-IP assays using the protein lysate prepared from YFP-Rab9–Tg mouse hearts, Rip1 and Ulk1 coimmunoprecipitated with YFP-Rab9, and the interaction was enhanced in the presence of starvation (Supplemental Figure 7H). We did not observe starvation-induced increases in the interaction between Rab9 and Rip1 in Ulk1-cKO mice. These results suggest that Ulk1, Rab9, and Rip1 form a complex in a Ulk1-dependent manner. Interestingly, syntaxin17, a newly proposed marker of autophagosomes (24), also interacted with YFP-Rab9 in a manner dependent on Ulk1 (Supplemental Figure 7I).
In order to demonstrate that Ulk1, Rab9, Rip1, and Drp1 form a single protein complex, heart homogenates prepared from mouse hearts were subjected to size-exclusion chromatography. Ulk1, Rip1, Drp1, and Rab9 were coeluted in fractions between 158 kDa and 670 kDa (fraction numbers 4, 5, and 6) at baseline and during starvation (Supplemental Figure 7J). Drp1 phosphorylated at S616 was eluted in fraction numbers 4 and 5 (Supplemental Figure 7J). Ulk1 and Rab9 were upregulated in fraction numbers 4 and 5 during starvation compared with baseline levels (Supplemental Figure 7K). We then repeated size fractionation of the heart homogenates prepared from YFP-Rab9–Tg mice and conducted co-IP assays using fraction numbers 4, 5, and 6. Ulk1, Rip1, and Drp1 were coimmunoprecipitated with YFP-Rab9 in fraction number 5 during starvation in a Ulk1-dependent manner (Figure 7D). These results suggest that Ulk1, Rab9, Rip1, and Drp1 form a protein complex in a Ulk1-dependent manner.
Next, we sought the intracellular location of the formation of this protein complex. Both Rip1 and Rab9 were found in the mitochondrial fraction in WT, Atg7-cKO, and Ulk1-cKO hearts at baseline (Supplemental Figure 8A). Their abundance in the mitochondrial fraction was not affected by starvation in any of the 3 groups. Using proximity ligation assays, we found that Rip1 interacted with YFP-Rab9 adjacent to mitochondria at baseline and that the interaction was enhanced during GD (Figure 8A). These data suggest that Rip1 interacts with Rab9 in the proximity of mitochondria, a process that is accelerated during GD and may be involved in the phosphorylation of Drp1 at S616.
 Figure 8
Figure 8Pharmacological inhibition of Rip1 impairs mitophagy during starvation in the heart. (A) CMs transduced with DsRed-Mito (green) were stained with rabbit anti-Rip1 antibody and/or mouse anti-GFP and anti-YFP antibodies, and in vivo protein-protein interaction (red dots) between Rip1 and YFP-Rab9 was detected with secondary proximity probes, Anti-Rabbit-PLUS, and Anti-Mouse MINUS using the Duolink In Situ PLA Detection Kit. Scale bars: 10 μm; original magnification, ×5 (enlarged insets). Arrows indicate the PLA signal close to the mitochondria. (B) Representative immunoblots showing phosphorylation of Drp1 in the presence or absence of 7-Cl-O-Nec-1 during glucose deprivation (GD) in CMs. (C) WT/Mito-Keima–Tg mice were subjected to 48 hours of starvation. The effect of 7-Cl-O-Nec-1 on mitophagy in the heart was examined. Either control inhibitor or 7-Cl-O-Nec-1 (1.80 μg/g BW) was injected i.p. twice during the 48-hour starvation period. Representative images and a summary are shown (n = 3 per group). Scale bars: 50 μm; original magnification, ×3 (enlarged insets). Error bars represent the SEM. #P < 0.05 versus control (unpaired Student’s t test). Nec-1, 7-Cl-O-Nec-1.
Consistent with the results above, treatment with 7-Cl-O-Nec-1, a specific inhibitor of Rip1 (25), suppressed the phosphorylation of Drp1 at S616 in a dose-dependent manner during GD in CMs (Figure 8B), suggesting that Rip1 mediates S616 phosphorylation of Drp1 in response to GD. Although Rip1 is a classical activator of NF-κB signaling (26), inhibition of NF-κB by forced expression of a phosphorylation-resistant mutant of IκBα did not affect the S616 phosphorylation of Drp1 during GD, suggesting that NF-κB activation is not required for the phosphorylation of Drp1 in response to energy stress (Supplemental Figure 8B). We found that 7-Cl-O-Nec-1 significantly decreased the area of high-ratio Mito-Keima dots induced by either energy stress or overexpression of Ulk1 (Supplemental Figure 8, C–F). These results suggest that Rip1 induces mitophagy through phosphorylation of Drp1 at S616 during energy stress in CMs. Intraperitoneal injection of necrostatin-1, an inhibitor of Rip1, the efficacy of which has been demonstrated in vivo (27), enlarged the average area of individual mitochondria and suppressed mitophagy in response to starvation in the hearts of WT mice (Supplemental Figure 8, G and H). However, the effect of nectrostatin-1 on mitochondrial size was no longer apparent in the hearts of Ulk1-cKO mice (Supplemental Figure 8, G and H). Consistently, we observed significantly fewer dots with high ratios of Mito-Keima signal during starvation in Mito-Keima–Tg mice treated with 7-Cl-O-Nec-1 than in those treated with control inhibitor (Figure 8C). These results indicate that Rip1 promotes mitochondrial fission and autophagy in a Ulk1-dependent manner during energy stress in the heart. Since Rip1 may also act as a pronecroptotic kinase, we evaluated the effect of 7-Cl-O-Nec-1 on ischemia-induced myocardial injury (Supplemental Figure 8, I and J). We found that 7-Cl-O-Nec-1 did not significantly affect the infarct size. This is consistent with the notion that Rip1 has both protective and detrimental functions that are mediated by mitophagy and necroptosis, respectively.
Pink1-Parkin–dependent mitophagy primarily targets depolarized mitochondria (4). Drp1-mediated mitochondrial fission also plays a role in segregating depolarized mitochondria (28). Therefore, we next asked whether Ulk1-Rab9–dependent mitophagy selectively degrades depolarized mitochondria. To this end, we evaluated the mitochondrial membrane potential with TMRE and tracked all mitochondria with MitoTracker Deep Red in CMs transduced with YFP-Rab9. At baseline, most mitochondria had a tubular morphology and high-intensity TMRE staining and appeared pink in the merged image (Supplemental Figure 8K), indicating that the mitochondrial membrane potential was maintained. YFP-Rab9 did not encircle these mitochondria (Supplemental Figure 8K). On the other hand, during GD, most mitochondria exhibited lower-intensity TMRE staining and appeared purple in the merged image (Supplemental Figure 8K). These mitochondria were fragmented and encircled by a YFP ring structure (Supplemental Figure 8K, yellow arrowheads). We observed many more purple dots encircled by YFP rings than pink dots, suggesting that Rab9-positive autophagosomes preferentially engulf depolarized mitochondria during GD. It should be noted that YFP-Rab9 did not interact with either Parkin or Nix, which are proteins known to interact with damaged mitochondria (Supplemental Figure 7F). These results suggest that Ulk1-Rab9–dependent mitophagy targets depolarized mitochondria using a mechanism distinct from that of Pink1-Parkin–dependent mitophagy.
S179 phosphorylation of Rab9 plays an essential role in mediating the assembly of the Ulk1-Rab9-Rip1-Drp1 complex and activating mitophagy in the heart. We further investigated the molecular mechanism through which this protein complex is formed. Ulk1 is phosphorylated by AMPK at multiple serine/threonine residues and is activated in response to energy stress (29, 30). We found that phosphorylation of Ulk1 at S555 and S317 was increased, whereas phosphorylation at S757 was downregulated during starvation (Supplemental Figure 9A). Of the multiple phosphorylation sites, S555 is largely preserved across species (30). In order to elucidate the role of Ulk1 phosphorylation at S555, we generated Ulk1 (S555A), a Ulk1 mutant in which S555 cannot be phosphorylated (Supplemental Figure 9B).
Co-IP assays revealed that both WT Ulk1 and a kinase-inactive mutant (K46N) of Ulk1 interacted with Rab9, whereas Ulk1 (S555A) did not (Supplemental Figure 9C). This suggests that phosphorylation of Ulk1 at S555, but not its kinase activity, is essential for the interaction between Ulk1 and Rab9.
Overexpression of Ulk1 (S555A) inhibited GD-induced increases in mitophagy, as evaluated with Mito-Keima (Supplemental Figure 9, D and E), suggesting that S555 phosphorylation of Ulk1 plays an essential role in mediating mitophagy during GD. In contrast, the level of LC3-II in CMs transduced with Ad-Ulk1-S555A was similar to that in Ad-LacZ–transduced CMs at baseline and during GD (Supplemental Figure 9F). Taken together, these data indicate that energy stress–induced phosphorylation of Ulk1 at S555 is essential for the recruitment of activated Rab9 during GD, which in turn mediates alternative autophagy. On the other hand, phosphorylation of Ulk1 at S555 does not appear to be essential for conventional autophagy.
Interestingly, overexpression of Ulk1 (K46N) inhibited the increase in the area of high-ratio Mito-Keima dots in response to GD (Supplemental Figure 9, D and E), even though the interaction of Ulk1 with Rab9 was preserved (Supplemental Figure 9C). Thus, although Ulk1 kinase activity is not required for the interaction between Ulk1 and Rab9, it is nevertheless required for mitophagy in CMs.
One possibility is that Ulk1 directly affects the GTPase activity of Rab9. The GTP-bound form of Rab mediates vesicular transport through specific interactions with effector molecules. Mannose-6-phosphate receptor–binding protein 1 (M6PRBP1) is a Rab9-binding protein that is required for endosome-to-Golgi transportation (31). Interaction with Rab9 increases the affinity of M6PRBP1 for its cargo. M6PRBP1-bound Rab9 was increased during GD (Supplemental Figure 9G), and knockdown of Ulk1 did not affect the increased binding of Rab9 to M6PRBP1 (Supplemental Figure 9G). These results suggest that Rab9 is activated in response to GD and that GD-induced activation of Rab9 occurs independently of Ulk1.
Given these observations, we hypothesized that Ulk1 directly phosphorylates Rab9 and that this phosphorylation plays an essential role in mediating mitophagy. To test this hypothesis, we analyzed the phosphorylation of Rab9 using Phos-tag SDS-PAGE. GD induced phosphorylation of Rab9 in CMs, which was inhibited in the presence of sh-Ulk1 but not 7-Cl-O-Nec-1 (Figure 9A). In vitro kinase assays revealed that recombinant Ulk1 directly phosphorylated Rab9 in a test tube (Supplemental Figure 9H). To identify the site at which Rab9 is phosphorylated by Ulk1, we performed mass spectrometric analyses. Tandem mass spectrometry (MS/MS) showed multiple peptide sequences modified by phosphorylation. Accordingly, we examined the relative abundance of phosphopeptide using Skyline software. This quantitative analysis revealed that S179 in the recombinant protein of human Rab9 purified from HEK293 cells was up to 99.92% phosphorylated by Ulk1 in a test tube (Supplemental Figure 9, I and J). This serine residue in Rab9 is conserved in multiple species (Supplemental Figure 9K) and is located in the complementary determining region 3 (CDR3). Consistent with the result obtained in CMs in vitro, phosphorylation of Rab9 was upregulated during starvation in WT but not Ulk1-cKO hearts (Supplemental Figure 9L).
 Figure 9
Figure 9Ulk1 directly phosphorylates Rab9 at S179, inducing mitochondrial fission and mitophagy. (A) Phosphorylation of Rab9 in CMs was analyzed with Phos-tag SDS-PAGE. A band of approximately 20 kDa in all 4 lanes represents an unphosphorylated form of Rab9. In lanes 2 and 4, anti-Rab9 antibody recognized a band with a higher molecular weight representing a monophosphorylated form of Rab9. (B) Phosphorylation of YFP-Rab9 in CMs was analyzed with Phos-tag SDS-PAGE. Anti-Rab9 antibody detected 2 bands in lane 2. The lower band corresponds to an unphosphorylated form of YFP-Rab9. The upper band in lane 2 was abolished in lane 3, suggesting that S179 is the unique phosphorylation site in Rab9. (C) Co-IP assays. (D and E) Representative immunoblots are shown. In D, Rab9 was detected on a parallel gel using the same samples.
To evaluate the functional role of Rab9 phosphorylation at S179, we transduced CMs with YFP-Rab9 (WT) or YFP-Rab9 (S179A), a phosphorylation-resistant mutant. Although YFP-Rab9 (WT) displayed a band shift indicative of phosphorylation on Phos-tag SDS-PAGE, YFP-Rab9 (S179A) did not (Figure 9B), suggesting that S179 is the major site of phosphorylation of Rab9 in CMs. We found that protein-protein interaction between YFP-Rab9 and Rip1 was impaired in the presence of YFP-Rab9 (S179A) (Figure 9C), suggesting that Ulk1 promotes Rab9-Rip1 interaction through phosphorylation of Rab9 at S179. Overexpression of Rab9 (S179A) suppressed the S616 phosphorylation of Drp1 during GD (Figure 9D), whereas Rab9 (S179D), a phosphorylation-mimetic mutant, induced S616 phosphorylation of Drp1 in a Rip1-dependent manner (Figure 9E). Unlike YFP-Rab9 (WT) (Figure 4B, left), YFP-Rab9 (S179A) did not encircle mitochondria during GD in the presence of chloroquine (Figure 4B, right, and Supplemental Figure 9M). Furthermore, CMs expressing YFP-Rab9 (WT) had round mitochondria (Figure 4B, left), whereas CMs expressing YFP-Rab9 (S179A) had more tubular mitochondria (Figure 4B, right, and Supplemental Figure 9M). Consistent with these results, forced expression of Rab9 (S179A) significantly suppressed the high-ratio Mito-Keima dots during energy stress, whereas Rab9 (S179D) significantly upregulated the high-ratio dots in a Rip1- and Drp1-dependent, but Parkin-independent, manner (Figure 10, A and B, and Supplemental Figure 10, A–C). Rab9 (S179A) significantly impaired ATP production in mitochondria isolated from CMs during GD compared with LacZ, suggesting that Rab9 phosphorylation at S179 plays an important role in maintaining mitochondrial activity during energy stress (Supplemental Figure 10D).
 Figure 10
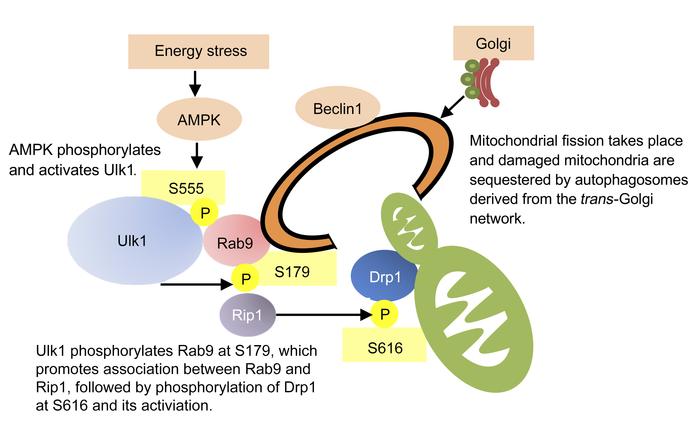
Figure 10The significance of phosphorylation of Rab9 at S179 in CMs in vitro. The effect of overexpression of mutant forms of Rab9 upon mitophagy was examined in CMs transduced with Mito-Keima. (A) Representative images. Scale bars: 50 μm; original magnification, ×2.8 (enlarged insets). (B) Quantitation of the area of high-ratio dots per total cell area. Number of cells in each group: 79, 16, and 20 (LacZ) and 25, 31, and 35 (Rab9 [S179A]) from 3 independent experiments. Error bars represent the SD. #P < 0.01 versus LacZ and no treatment; †P < 0.01 versus LacZ and GD; *P < 0.01 versus LacZ and hypoxia (Tukey-Kramer’s test). (C) Proposed model of mitophagy in response to energy stress.
In summary, our results suggest that Ulk1-dependent phosphorylation of Rab9 at S179 plays an essential role in mediating energy stress–induced mitophagy by facilitating interaction between Rab9 and Rip1 and the consequent phosphorylation of Drp1 at S616. We propose that S555-phosphorylated Ulk1 acts as a scaffold to assemble a complex comprising Rab9, presumably associated with trans-Golgi membranes, and the Rip1-Drp1 complex, so that mitochondria labeled with S616-phoshorylated Drp1 are sequestrated by phagophores assembled via Rab9 (Figure 10C).
Mitophagy in response to energy stress in the heart is selectively abolished in Rab9 (S179A) knockin mice. We hypothesized that suppression of Rab9 phosphorylation at S179 may selectively abolish energy stress-induced mitophagy, without affecting general autophagy in the heart in vivo. This would be in contrast to the effect of Atg7-cKO hearts, in which general autophagy, but not mitophagy, is abolished. We generated knockin (KI) mice expressing phosphorylation-resistant Rab9 (S179A) and evaluated mitophagy in the heart. This KI mouse, which was crossed with the GFP-LC3–Tg mouse, showed preserved LC3-dependent autophagic flux in the heart during starvation (Supplemental Figure 11, A and B). Consistent with the results obtained in vitro, mitophagy was significantly impaired in the heart of the Rab9 (S179A) KI (Rab9-KI) mouse during ischemia (Figure 11, A and B), demonstrating the relevance of this phosphorylation in vivo. Mitochondria isolated from the hearts of Rab9-KI or Ulk1-cKO mice displayed reduced ATP production and complex I and IV activity during ischemia compared with those from control mice (Supplemental Figure 11, C and D). Following 2 hours of ischemia, the infarct size/AAR was significantly greater in Rab9-KI mice than in WT mice (Figure 11, C and D). These results suggest that phosphorylation of Rab9 at S179 mediates mitophagy and plays an essential role in protecting the heart against ischemic injury. Furthermore, our findings indicate that mitophagy mediated by the Ulk1-Rab9-Rip1-Ulk1–dependent mechanism plays an essential role in protecting the heart in response to energy stress.
 Figure 11
Figure 11Significance of phosphorylation of Rab9 at S179 in the heart. (A and B) Lysosomal degradation of mitochondria was examined in the hearts of Mito-Keima–Tg mice during ischemia. Representative images are shown in A. Scale bars: 50 μm; original magnification, ×3 (enlarged insets). A summary is shown in B (n = 3 per group). #P < 0.01 versus WT and no ischemia; ‡P < 0.01 versus WT and ischemia (Tukey-Kramer’s test). (C) Representative images of LV slices with Alcian blue and TTC staining. Scale bars: 5 mm. (D) AAR/LV and infarct size/AAR. n = 7–8 per group. #P < 0.01 versus WT (unpaired Student’s t test). Error bars represent the SEM.
Mitophagy is activated by the Ulk1-Rab9–dependent mechanism in human CMs. We evaluated whether the Ulk1-Rab9–dependent form of mitophagy is relevant to human heart disease. We first tested whether mitophagy during energy stress is mediated through the Rab9-dependent mechanism in CMs derived from human induced pluripotent stem cells (hiPSC-CMs) (32). To this end, hiPSC-CMs were subjected to GD, and mitophagy was observed with Mito-Keima. As with the results obtained from rodent CMs, mitophagy was induced by GD in hiPSC-CMs, and the induction of mitophagy was significantly suppressed in the presence of Rab9 (S179A) (Supplemental Figure 11E), indicating that mitophagy is induced through the Rab9 S179 phosphorylation–dependent mechanism in hiPSC-CMs. We have shown previously that the level of mitophagy is insufficient to maintain mitochondrial function in decompensated hearts (14). Phosphorylation of Ulk1 at S555 and Drp1 at S616 was downregulated in human hearts obtained from patients with end-stage dilated cardiomyopathy (DCM) compared with that seen in hearts from donors with normal cardiac function (Supplemental Figure 11F and Supplemental Table 1), suggesting that the Rab9-dependent form of mitophagy may be downregulated in patients with heart failure.
Finally, we examined whether Ulk1-Rab9–dependent mitophagy can degrade other organelles. Since ER-specific autophagy, or ERphagy, was recently reported in mammalian cells (33), we examined the ER content in our mouse model. We found that the ER protein level was not significantly affected at baseline or during starvation in WT, Atg7-cKO, or Ulk1-cKO mice (Supplemental Figure 11G). Thus, whether the Ulk1-Rab9-dependent mitophagy degrades ER remains to be elucidated.
-
Discussion
We believe the present study provides compelling evidence that autophagy characterized by Ulk1-dependent activation and Rab9-positive autophagosomes, distinct from that characterized by the requirement of the Atg conjugation system and LC3-postive autophagosomes (18), predominantly mediates mitophagy and is chiefly responsible for the maintenance of mitochondrial quality in the heart during energy stress. Energy stress in the heart induces the formation of a multiprotein complex consisting of Ulk1, Rab9, Rip1, and Drp1, which allows the recruitment of late endosome membranes associated with Rab9 to damaged mitochondria through the coordinated actions of Rab9 phosphorylated by Ulk1 at S179 and Drp1 phosphorylated by Rip1 at S616. In Rab9-KI mice, ischemia-induced mitophagy activation was selectively attenuated, while induction of general autophagy was unaffected, providing clear evidence that mitophagy and general autophagy are mediated by distinct mechanisms in vivo. Furthermore, these mice exhibited more severe mitochondrial dysfunction and myocardial damage than did control mice, suggesting that mitophagy mediated by the Ulk1-Rab9-Rip1-Drp1–dependent mechanism is critically involved in cardioprotection during myocardial ischemia.
Activation of mitophagy during energy stress is mediated by Ulk1- and Rab9-dependent mechanisms in the heart. Increasing lines of evidence suggest that nonconventional autophagy can degrade mitochondria in mammalian cells. However, the molecular constituents of autophagosomes without LC3 have remained obscure. Using Immunogold EM, we observed that autophagosomes harboring mitochondria associated with Rab9, but not LC3, in a Ulk1-dependent manner. Furthermore, we found that Rab9-YFP–positive ring-like structures with a diameter of 500 to 1500 nm, comparable to the size of autophagosomes, were colocalized with neutral Mito-Keima dots in CMs subjected to GD and that some of the ring-like structures surrounded acidic Mito-Keima dots. These results suggest that Rab9-positive autophagosomes contain mitochondria in CMs subjected to energy stress and that they are transferred to lysosomes to become autolysosomes. Rab9 is involved in transport between late endosomes and the trans-Golgi network (34). Together with the fact that mitophagy induced by energy stress is inhibited in the presence of BFA, these results strongly suggest that mitochondria are engulfed by Rab9-positive phagophores that are probably derived from the late endosomes and the trans-Golgi network, a process similar to the cargo engulfment seen in alternative autophagy (18). Genetic evidence suggests that Rab family proteins are essential mediators of mitophagy in yeast (35).
Recent evidence suggests that damaged mitochondria can be degraded by mitochondria-derived vesicles (MDVs) (36) and a Rab5-dependent endosomal pathway (37). The mitophagy described in this work is distinct from those mechanisms, in that neither MDVs nor the endosomal pathway utilizes Drp1. Moreover, although both MDVs and the endosomal pathway are Parkin dependent, mitophagy by alternative autophagy is Parkin independent. In addition, the endosomal pathway is Ulk1 independent. Whether MDVs and the endosomal pathway also contribute to the clearance of damaged mitochondria during energy stress in the heart in vivo and, if so, to what extent they contribute to mitochondrial quality control remains to be elucidated.
Mitophagy mediated through alternative autophagy is Drp1 dependent. We and others have shown previously that Drp1 plays an essential role in mediating mitophagy in the heart and that this form of mitophagy is unique in that it is Parkin independent (9, 14). We show here that mitophagy mediated through the Ulk1-dependent mechanism is abolished in the absence of Drp1 (Figure 6). A genetic screen showed that DNM1, a homolog of Drp1, is an essential component of mitophagy in yeast (35), in which the scaffold protein ATG11 interacts with DNM1, so that the fission complex associates with mitochondria undergoing degradation. Mitochondrial fission is also believed to be an important process for segregating depolarized mitochondria from healthy ones before mitophagy ensues in mammalian cells (28). Alternatively, mitochondrial fission may be needed to protect the healthy part of mitochondria from mitophagic degradation (38). However, how Drp1 coordinates with the autophagic machinery to mediate mitophagy has not been elucidated. Recent evidence suggests that segregation of damaged sections of mitochondria from the healthy sections can take place independently of Drp1, possibly by physical force exerted by an extended phagophore membrane, in HeLa cells, SH-SY5Y cells, and MEF cells (39). As discussed below, however, we propose that Drp1 is an integral component of the core machinery responsible for mitophagy in CMs.
Mitophagy is mediated by coordinated actions of Ulk1, Rab9, Rip1, and Drp1 in CMs. Although previous studies have indicated that a special form of general autophagy, such as that referred to as alternative autophagy, is activated in both a Ulk1- and Rab9-dependent manner and that it can degrade mitochondria (10), the molecular mechanisms by which autophagosomes target mitochondria are not clearly understood. We propose that a protein complex, consisting of Ulk1, Rab9, Rip1, and Drp1, is formed in a manner that is dependent on phosphorylation of Ulk1 at S555 and of Rab9 at S179 and acts as a cellular assembly site for autophagosomes with Rab9-associated intracellular membranes and mitochondria marked by Drp1.
First, energy stress activates AMPK, which in turn phosphorylates Ulk1 at multiple serine/threonine residues, including S555 (30). Although phosphorylation of 4 serine residues, including S555, in Ulk1 abolishes conventional autophagy in mouse embryonic fibroblasts (MEFs) (30), we found that interaction of Ulk1 with Rab9 is dependent on phosphorylation of S555 and that S555A mutation selectively abolishes mitophagy without affecting conventional autophagy in response to GD in CMs. Interestingly, interaction between Ulk1 and Rab9 does not require the kinase activity of Ulk1. Thus, Ulk1 appears to function solely as a scaffold at this stage, rather than as a kinase, recruiting Rab9 carrying trans-Golgi membranes. How intracellular organelle membranes recruited by GTP-loaded Rab9 develop into autophagosomes remains to be clarified. For example, whether the ubiquitin-like conjugation system is required in this process is currently unknown.
In the second step, Ulk1 promotes the association of Rab9 with Rip1, a Drp1-kinase, which in turn phosphorylates Drp1 at S616. This phosphorylation is known to promote mitochondrial translocation of Drp1 and fission. Importantly, Ulk1 does not phosphorylate Drp1 directly but rather promotes interaction between Rip1 and Drp1 through S179 phosphorylation of Rab9, and Rip1 in turn phosphorylates Drp1 at S616. Rip1 is known to mediate death receptor–induced necroptosis. However, increasing lines of evidence suggest that it also participates in mitochondrial events, including fission, as a physiological kinase (20). Here, we show that both Rip1 and Drp1 play an essential role in mediating mitophagy in CMs in response to energy stress. We speculate that the association of Drp1 with the Ulk1-Rab9 complex allows S616-phosphorylated Drp1 to present damaged mitochondria, after segregation by fission, for recognition by phagophores derived from intracellular membranes associated with GTP-loaded Rab9 generated on the Ulk1-Rab9 platform. Since Ulk1, Rab9, Rip1, and Drp1 coexist in a large molecular-weight complex, other molecules may also coexist in this complex to facilitate mitophagy. This form of mitophagy preferentially sequestrates depolarized mitochondria in CMs subjected to GD. Whether depolarization of mitochondrial membrane potential triggers association of an unknown mitophagy receptor with the Ulk1-Rab9-Rip1-Drp1 complex remains to be clarified.
General autophagy and mitophagy may be regulated by distinct cellular mechanisms in vivo. We have previously shown that mitophagy is activated in the heart subjected to pressure overload after conventional autophagy is inactivated below physiological levels (14). This, together with the current results, suggests that mitophagy and general autophagy can be regulated by distinct mechanisms in the heart and that Rab9-KI mice are useful to elucidate the role of mitophagy in the heart under stress. Since mitochondrial dysfunction and downregulation of mitophagy are observed in many pathological conditions in the heart, exploring mechanisms uniquely involved in mitophagy may allow the development of novel strategies to improve the quality of mitochondria in the heart under stress, even when general autophagy is downregulated.
-
Methods
Animal models. WT, Atg7-cKO, Ulk1-cKO, Parkin-KO, and Rab9-KI mice (2 to 3 months of age) were used (C57BL/6 background). The genetic background of the Mito-Keima–Tg, YFP-Rab9–Tg, and GFP-LC3–Tg mice (Myh6-driven transgene) was adjusted by crossing these mice with C57BL/6 inbred mice for more than 5 generations. The Atg7-floxed mice were a gift of Masaaki Komatsu (Niigata University, Niigata, Japan). The Ulk1-floxed mice were a gift of Mondira Kundu (St. Jude Children’s Research Hospital, Memphis, Tennessee, USA). The Myh6-Cre mice, a gift of Michael D. Schneider (Imperial College London, London, United Kingdom), were used to generate cardiac-specific KO. The Parkin-KO mice (B6.129S4-Park2tm1Shn/J) were purchased from The Jackson Laboratory. The Rab9-KI mice were generated by the Genome Editing Shared Resource at Rutgers University. Pathogen-free mice were housed in a temperature-controlled environment with 12-hour light/12-hour dark cycles and were given food and water ad libitum.
Starvation of mice. To induce autophagy throughout the body, mice were starved for 48 hours (15). During starvation, the animals had free access to water.
Ischemia surgery. Mice were randomly divided into 2 groups — sham or ischemia surgery — and were anesthetized by i.p. injection of pentobarbital sodium (60 mg/kg). A rodent ventilator was used with 65% oxygen for ventilation. The animals were maintained at a temperature ranging from 36.5°C to 37.5°C. The chest was opened by a horizontal incision through the muscle between the ribs (third intercostal space). Ischemia was achieved by ligating the anterior descending branch of the left anterior descending artery (LAD) using an 8-0 nylon suture with silicon tubing (1 mm OD) placed on top of the LAD, 2 mm below the border between the left atrium and left ventricle (LV). Regional ischemia was confirmed by electrocardiographic (ECG) change (ST elevation) (15). All surgeries were conducted in a blinded manner with respect to mouse genotype. The ischemic zone in histochemical and biochemical assays was analyzed.
Assessment of AAR and infarct size. After 2 hours of ischemia, the animals were reanesthetized and intubated and their chests opened. After arresting the heart at the diastolic phase by KCl injection, the ascending aorta was cannulated and perfused with saline to wash out blood. To demarcate the ischemic AAR, Alcian blue dye (1%) was perfused into the aorta and coronary arteries. The hearts were excised and sliced into 1-mm-thick cross sections. The slices were then incubated with a 1% triphenyltetrazolium chloride (TTC) solution at 37°C for 10 minutes to evaluate infarct size (16). All processes were conducted in a blinded manner with respect to mouse genotype.
CM cultures. Primary cultures of neonatal rat ventricular CMs were prepared from 1-day-old Crl:(WI) BR-Wistar rats (Charles River Laboratories). A CM-rich fraction was obtained and maintained in culture as described previously (15). hiPSCs were differentiated into beating hiPSC-CMs as described previously (32).
GD. CMs were washed 3 times with PBS and incubated for 4 hours in glucose-free, serum-free DMEM (Invitrogen, Thermo Fisher Scientific) as described previously (15).
Hypoxia. Hypoxic conditions were created by incubating the cells in a Modular Incubator Chamber (Billups-Rothenberg Inc.) in an atmosphere of 5% CO2, 95% N2, and 37°C for 4 hours.
Chemical inhibitors. To inhibit autophagic flux in vivo, chloroquine was injected i.p. (10 μg/kg). Four hours later, the animals were euthanized for the detection of autophagy markers. To inhibit autophagic flux in vitro, cultured CMs were treated with 10 μM chloroquine (Fluka Biochemika) for 4 hours or with 50 nM bafilomycin A1 (MilliporeSigma) for 2 hours as described previously (15). To inhibit trafficking of membranes derived from Golgi in cultured CMs, BFA (Cell Signaling Technology) was applied for 3 hours during GD at the concentrations indicated in Supplemental Figure 3, G, H, J, and K (18). To inhibit the kinase activity of Rip1 in cultured CMs, 7-Cl-O-Nec-1 (EMD Millipore) was applied for 3 hours during GD at the concentrations indicated in Figure 8B, Figure 9E, Supplemental Figure 8, E and F, and Supplemental Figure 10, A and B (25). To inhibit the kinase activity of Rip1 in vivo, nectrostatin-1 (EMD Millipore) (1.65 μg/g BW) or 7-Cl-O-Nec-1 (1.80 μg/g BW) was injected i.p. twice during the 48-hour starvation period (27). Necrostatin-1, Inactive Control (EMD Millipore) was used as a control.
Adenoviral constructs and transduction. CMs were transduced with adenovirus for overexpression for 48 hours and for gene silencing for 96 hours. A MOI of 3 to 5 was used for overexpression and a MOI of 10 to 15 was used for gene silencing. Adenoviruses harboring shRNA against Atg7 (Ad-sh-Atg7), Ulk1 (Ad-sh-Ulk1), Beclin1 (Ad-sh-Beclin1), Drp1 (Ad-sh-Drp1), Rab9 (Ad-sh-Rab9), and Parkin (Ad-sh-Parkin) were generated as previously described (15) using the following hairpin-forming oligonucleotides: 5′-CGGTCAAAGGACAAAGTTAACATTCAAGAGATGTTAACTTTGTCCTTTGACCTTTTTA-3′ for Ad-sh-Atg7; 5′-CGACTTCCAGGAGATGGCCAATTCAAGAGAATTGGCCATCTCCTGGAAGTCTTTTTA-3′ for Ad-sh-Ulk1; 5′-CAATTTGGCACGATCAATATTCAAGAGATATTGATCGTGCCAAATTGCTTTTTTTT-3′ for Ad-sh-Beclin1; 5′-CGGCAATTGAGCTAGCATATTTCAAGAGAATATGCTAGCTCAATGCCTTTTTA-3′ for Ad-sh-Drp1; 5′-CGGGCAACAAGACTGACATAATTCAAGAGATTATGTCAGTCTTGTTGCCCTTTTTA-3′ for Ad-sh-Rab9; and 5′-CGCCGCACGACCTCATGGGAATTCAAGAGATTCCCATGAGGTCGTGCGGC TTTTTA-3′ for Ad-sh-Parkin.
Evaluation of fluorescent LC3 puncta. For observation of LC3 puncta in the heart, GFP-LC3–Tg mice were used as described previously (15). Hearts were excised, washed with PBS, and fixed with 4% paraformaldehyde (PFA) (pH 9.5) for 30 minutes. Immediately after the fix, the heart sections were prepared using a vibratome and subjected to confocal imaging. Images were acquired by an unbiased observer blinded to mouse genotype and experimental conditions, such as baseline values, starvation, or chloroquine treatment.
EM analyses. CMs were fixed in Karnovsky’s fixative and then postfixed in 1% osmium tetraoxide, dehydrated in a graded series of acetone concentrations, and embedded in Sparr resin. Sections of 98-nm thickness were placed on copper grids that were double-stained with uranyl acetate and lead citrate. Discs were examined with a JEOL 1200 electron microscope. All procedures were conducted in a blinded manner with respect to the genotype of the mice.
Keima with mitochondrial targeting signal. Keima with mitochondrial localization signaling (Mito-Keima) acts as a mitochondrially localized pH indicator protein, as described by Katayama et al. (17). For observation of mitophagy in cultured CMs, adenovirus harboring Mito-Keima was generated (16). All images were acquired using living cells at 37°C by Nikon A1RSI confocal microscopy. For observation of mitophagy in the heart, Mito-Keima–Tg mice were generated. Hearts were excised, washed with PBS, and fixed with 4% PFA (pH 9.5) for 30 minutes. Immediately after the fixation, the heart sections were prepared by vibratome and subjected to confocal imaging. Images were acquired by an unbiased observer blinded to mouse genotype and experimental conditions, such as baseline values, starvation, or ischemia.
Immunohistochemistry. CMs cultured on coverslips were fixed with 4% PFA, permeabilized in PBS containing 0.2% Triton-X, and blocked with 3% BSA for 30 minutes, followed by incubation with a primary antibody. The primary antibodies used in this study were as follows: anti-Lamp2 (Abcam, ab37024), anti-Lamp2A (Abcam, ab18528), and anti-Rab9 (Abcam, ab2810). Alexa Fluor 488– or Alexa Fluor 594–conjugated secondary antibodies (Invitrogen, Thermo Fisher Scientific) were used for the detection of indirect fluorescence (15).
Quantitative real-time PCR for mitochondrial DNA. The mtDNA content was quantified by real-time PCR of cardiac DNA (16). Experiments were conducted in duplicate in order to exclude the possibility of technical variance. Primer sequences for cytochrome b and Actb were as follows: 5′-CCACTTCATCTTACCATTTATTATCGC-3′ (forward primer) and 5′-TTTTATCTGCATCTGAGTTTAA-3′ (reverse primer) for cytochrome b, and 5′-CTGCCTGACGGCCAGG-3′ (forward primer) and 5′-CTATGGCCTCAGGAGTTTTGTC-3′ (reverse primer) for genomic Actb.
Evaluation of mitochondrial membrane potential and integrity. In order to evaluate mitochondrial membrane potential and integrity, staining of cultured CMs with TMRE and JC-1 was conducted using MitoPT TMRE and MitoPT JC-1 (ImmunoChemistry Technologies), respectively, according to the manufacturer’s instructions.
Evaluation of ATP synthesis. Mitochondria were freshly isolated from the ventricle using a Mitochondria Isolation Kit (MilliporeSigma, MITOISO1). The mitochondria isolated from 50-mg tissue samples were suspended in 50 ml storage buffer (10 mM HEPES, pH 7.4, 250 mM sucrose, 2 mM K2HPO4), resulting in a final volume of approximately 100 ml of mitochondrial fraction. ATP production was measured by luminometric assay using an ATP Bioluminescence Assay Kit (MilliporeSigma, FL-AA), as described previously (40).
Evaluation of mitochondrial complex activity. Electron transport chain complex activities were measured using the MitoCheck Complex I and IV Activity Assay Kit (Cayman Chemical Company) according to the method described previously (16).
Evaluation of cell death. In order to evaluate cell death, staining of cultured CMs with Propidium Iodide Nucleic Acid Stain (Invitrogen, Thermo Fisher Scientific) was conducted according to the manufacturer’s instructions.
Immunoblot analysis. The primary antibodies used in this study were as follows: anti-LC3 (MBL, M186-3); anti-p62/SQSTM1 (Origene, TA307334); anti-Atg7 (Cell Signaling Technology, 8558); anti-Ulk1 (MilliporeSigma, A7481); anti-Parkin (Cell Signaling Technology, 2132); anti-GAPDH (Cell Signaling Technology, 2118); anti–α-tubulin (MilliporeSigma, T6199); anti–total OXPHOS rodent WB antibody cocktail (Abcam, ab110413); anti-Rab9 (Cell Signaling Technology, 5118); anti-Drp1 (BD Transduction, 611112); anti-Rip1 (Cell Signaling Technology, 3493); anti–phosphorylated Drp1 (anti–p-Drp1) at S616 (Cell Signaling Technology, 4494); anti–p-Ulk1 (S555) (Cell Signaling Technology, 5869); anti–p-Ulk1 (S317) (Cell Signaling Technology, 12753); anti–p-Ulk1 (S757) (Cell Signaling Technology, 6888); anti-CDK1 (Cell Signaling Technology, 77055); anti-CDK5 (Cell Signaling Technology, 14145); anti-ERK2 (Cell Signaling Technology, 9108); anti–syntaxin 17 (MilliporeSigma, HPA001204); and anti-HA (Cell Signaling Technology, 3724). The secondary antibodies used in this study were as follows: anti–rabbit IgG HRP-linked antibody (Cell Signaling Technology, 7074) and anti–mouse IgG HRP-linked antibody (Cell Signaling Technology, 7076).
IP. CMs were lysed with IGEPAL CA-630 buffer (50 mM Tris-HCl [pH 7.4], 1% IGEPAL CA-630, 10 mM EDTA, 150 mM NaCl, 50 mM NaF, 1 μM leupeptin, and 0.1 μM aprotinin) containing phosphatase inhibitor. For IP of YFP-Rab9, the GFP-Trap-A Kit (Chromotek) was used according to the manufacturer’s instructions. For IP of HA-Drp1, the Pierce HA Tag IP Kit (Thermo Fisher Scientific) was used according to the manufacturer’s instructions.
Size exclusion chromatography. Using the BioCAD SPRINT Perfusion Chromatography System (PerSpective BioSystems), clarified heart tissue lysate was injected (100 μl per injection) onto a TSKgel UltraSW Aggregate column (Tosoh Bioscience) equilibrated with 50 mM Tris and 150 mM NaCl (pH 7.5). The protein elution profile was detected by UV absorbance at 280 nm. To assess the size of the protein complex of interest, the column was calibrated with protein standards (Bio-Rad Laboratories) consisting of thyroglobulin (bovine) (670,000 Da), γ-globulin (bovine, 158,000 Da), OVA (chicken, 44,000 Da), myoglobin (horse, 17,000 Da), and vitamin B12 (1350 Da).
In situ proximity ligation assay. Cultured CMs were fixed, permeabilized, and blocked as described above. CMs were incubated with primary antibodies (rabbit anti-Rip1 and mouse anti-GFP/anti-YFP in blocking solution) at room temperature for 2 hours. After washing, the protein-protein interaction between Rip1 and YFP-Rab9 was detected with secondary proximity probes (Rabbit-PLUS and Mouse-MINUS, Olink Biosciences AB). The secondary proximity probes were incubated for 1 hour at 37°C. All subsequent steps were performed according to the Duolink proximity ligation assay (PLA) detection kit protocol (Olink Biosciences AB).
Phos-tag SDS-PAGE analysis of Rab9 phosphorylation. Protein samples were separated by SDS-PAGE on polyacrylamide gels containing 30 μM Phos-tag (VWR, Wako) and 60 μM MnCl2. After electrophoresis, the gel was soaked in transfer buffer (25 mM Tris, 192 mM glycine, 10% methanol) containing 2 mM EDTA for 40 minutes to remove Mn2+ and then in transfer buffer without EDTA for 20 minutes. Proteins were then transferred to PVDF membranes (Bio-Rad).
In vitro kinase assay. An in vitro kinase assay was carried out as described previously, with modifications (15). Recombinant active human Ulk1 protein fragment (10 ng) (Abcam) was incubated with recombinant human Rab9 protein (1 μg) (Origene) in a kinase buffer (50 mM HEPES [pH 7.4], 15 mM MgCl2 and 200 μM sodium vanadate containing 100 μM ATP or 5 μM ATP and 10 μCi [γ-32P] ATP per reaction) at 30°C for 30 minutes. Phosphorylated proteins were separated by SDS-PAGE and analyzed by autoradiography.
MS analysis. The protein gel band with the molecular weight corresponding to Rab9 was cut for in-gel trypsin digestion followed by phosphorylation site identification using liquid chromatography MS/MS (LC-MS/MS) analysis. In brief, the gel band was excised, followed by DTT reduction and iodoacetamide alkylation. Trypsin was used for protein digestion at 37°C overnight. The resulting peptides were desalted using a C18 Ziptip (EMD Millipore) prior to LC-MS/MS analysis. Peptide separation was performed on an Ultimate 3000 Nano UHPLC System (Thermo Fisher Scientific) using a C18 reversed-phase nano column (Acclaim PepMap RSLC, 75 μm × 50 cm, 2 μm, 100 Å, Thermo Fisher Scientific). The eluted peptides were directly introduced into a Q Exactive MS system, and the MS/MS spectra were acquired as data-dependent analyses, in which the top-15 intensity peptides were selected in MS full-scan mode for MS/MS analysis. MS/MS spectra were search against the SwissProt human database using the Mascot (version 2.3) search engine on the Proteome Discoverer (version 1.4) platform. Oxidized methionine, carbamidomethyl alkylated cysteine, and serine/threonine phosphorylation were set as variable modifications. The phosphorylation site assignment was estimated using the PhosphoRS node in the Proteome Discoverer software program and then confirmed by manual evaluation of the MS/MS spectra. The peak areas of phosphopeptides and nonphosphopeptides were acquired using Skyline software. The relative abundance of a phosphopeptide was calculated as follows: percentage of phosphopeptide = 100% × peak area of phosphopeptide/(peak area of phosphopeptide + peak area of nonphosphopeptide).
Human samples from explanted hearts. The heart samples used in this study were obtained from donors and patients who had undergone heart transplantation (Supplemental Table 1) at Taipei Veterans General Hospital. Myocardial left atrium samples were collected at the time of therapeutic heart transplantation. Immediately after tissue procurement, the samples were stored in liquid nitrogen and kept at –80°C.
Statistics. Data are expressed as the mean ± SEM or SD. A 2-tailed Student’s t test or 1-way ANOVA with Tukey-Kramer’s test was used to determine statistical significance of differences between 2 groups or among multiple groups, respectively. A P value of less than 0.05 was considered statistically significant.
Study approval. All animal protocols were approved by the IACUC of Rutgers University New Jersey Medical School. The study using human heart samples was approved by the ethics committee of Taipei Veterans General Hospital. All patients or their families provided written informed consent.
-
Author contributions
TS designed the study, performed most of the experiments, analyzed data, interpreted results, and wrote the manuscript. JN, SO, RM, Yasuhiro Maejima, Yoshiya Monden, YI, SS, EB, and CPH performed experiments and analyzed data. TL and HL performed proteomic experiments, analyzed data, and interpreted results. LF performed confocal microscopic analyses. PZ performed surgical manipulations of mice. SI performed EM analyses with Immunogold. DF and MI provided project resources. MK generated the Ulk1fl/fl mice and advised on the mechanism of mitophagy. JS designed and supervised the study, interpreted results, provided project resources, and wrote the manuscript. All authors reviewed and commented on the manuscript.
-
Acknowledgments
The authors thank Daniela Zablocki (Rutgers New Jersey Medical School) for critical reading of the manuscript. This work was supported in part by grants from the US Public Health Service (HL67724, HL91469, HL102738, HL112330, HL138720, and AG23039, to JS) and the Leducq Foundation Transatlantic Network of Excellence (15CBD04 to JS). TS was supported by a postdoctoral fellowship from the American Heart Association and a research fellowship from the Japan Heart Foundation/Bayer Yakuhin.
Address correspondence to: Junichi Sadoshima, Department of Cell Biology and Molecular Medicine, Cardiovascular Research Institute, Rutgers New Jersey Medical School, 185 S. Orange Avenue, MSB G609, Newark, New Jersey 07103, USA. Phone: 973.972.8916; Email: sadoshju@njms.rutgers.edu.
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(2):802–819.https://doi.org/10.1172/JCI122035.
See the related Commentary at Ulk1/Rab9-mediated alternative mitophagy confers cardioprotection during energy stress.
-
References
- Gatica D, Chiong M, Lavandero S, Klionsky DJ. Molecular mechanisms of autophagy in the cardiovascular system. Circ Res. 2015;116(3):456–467.
- Friedman JR, Nunnari J. Mitochondrial form and function. Nature. 2014;505(7483):335–343.
- Saito T, Sadoshima J. Molecular mechanisms of mitochondrial autophagy/mitophagy in the heart. Circ Res. 2015;116(8):1477–1490.
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183(5):795–803.
- Kane LA, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205(2):143–153.
- Koyano F, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510(7503):162–166.
- Chen Y, Dorn GW. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340(6131):471–475.
- Lazarou M, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524(7565):309–314.
- Kageyama Y, et al. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 2014;33(23):2798–2813.
- Hirota Y, et al. Mitophagy is primarily due to alternative autophagy and requires the MAPK1 and MAPK14 signaling pathways. Autophagy. 2015;11(2):332–343.
- Honda S, Arakawa S, Nishida Y, Yamaguchi H, Ishii E, Shimizu S. Ulk1-mediated Atg5-independent macroautophagy mediates elimination of mitochondria from embryonic reticulocytes. Nat Commun. 2014;5:4004. View this article via: PubMed Google Scholar
- Hom J, Sheu SS. Morphological dynamics of mitochondria--a special emphasis on cardiac muscle cells. J Mol Cell Cardiol. 2009;46(6):811–820.
- Lam MP, et al. Protein kinetic signatures of the remodeling heart following isoproterenol stimulation. J Clin Invest. 2014;124(4):1734–1744.
- Shirakabe A, et al. Drp1-dependent mitochondrial autophagy plays a protective role against pressure overload-induced mitochondrial dysfunction and heart failure. Circulation. 2016;133(13):1249–1263.
- Maejima Y, et al. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat Med. 2013;19(11):1478–1488.
- Ikeda Y, et al. Endogenous Drp1 mediates mitochondrial autophagy and protects the heart against energy stress. Circ Res. 2015;116(2):264–278.
- Katayama H, Kogure T, Mizushima N, Yoshimori T, Miyawaki A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol. 2011;18(8):1042–1052.
- Nishida Y, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461(7264):654–658.
- Mao K, Wang K, Liu X, Klionsky DJ. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev Cell. 2013;26(1):9–18.
- Wang X, et al. RNA viruses promote activation of the NLRP3 inflammasome through a RIP1-RIP3-DRP1 signaling pathway. Nat Immunol. 2014;15(12):1126–1133.
- Kashatus JA, et al. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol Cell. 2015;57(3):537–551.
- Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282(15):11521–11529.
- Jahani-Asl A, et al. CDK5 phosphorylates DRP1 and drives mitochondrial defects in NMDA-induced neuronal death. Hum Mol Genet. 2015;24(16):4573–4583.
- Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151(6):1256–1269.
- Degterev A, Zhou W, Maki JL, Yuan J. Assays for necroptosis and activity of RIP kinases. Meth Enzymol. 2014;545:1–33.View this article via: PubMed Google Scholar
- Regula KM, Ens K, Kirshenbaum LA. IKK beta is required for Bcl-2-mediated NF-kappa B activation in ventricular myocytes. J Biol Chem. 2002;277(41):38676–38682.
- Lim SY, Davidson SM, Mocanu MM, Yellon DM, Smith CC. The cardioprotective effect of necrostatin requires the cyclophilin-D component of the mitochondrial permeability transition pore. Cardiovasc Drugs Ther. 2007;21(6):467–469.
- Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27(2):433–446.
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141.
- Egan DF, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331(6016):456–461.
- Carroll KS, Hanna J, Simon I, Krise J, Barbero P, Pfeffer SR. Role of Rab9 GTPase in facilitating receptor recruitment by TIP47. Science. 2001;292(5520):1373–1376.
- Baljinnyam E, et al. Effect of densely ionizing radiation on cardiomyocyte differentiation from human-induced pluripotent stem cells. Physiol Rep. 2017;5(15):e13308. View this article via: PubMed Google Scholar
- Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson ÅB. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. 2012;287(23):19094–19104.
- Lombardi D, Soldati T, Riederer MA, Goda Y, Zerial M, Pfeffer SR. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 1993;12(2):677–682.
- Kanki T, et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell. 2009;20(22):4730–4738.
- Sugiura A, McLelland GL, Fon EA, McBride HM. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 2014;33(19):2142–2156.
- Hammerling BC, et al. A Rab5 endosomal pathway mediates Parkin-dependent mitochondrial clearance. Nat Commun. 2017;8:14050. View this article via: PubMed Google Scholar
- Burman JL, et al. Mitochondrial fission facilitates the selective mitophagy of protein aggregates. J Cell Biol. 2017;216(10):3231–3247.
- Yamashita SI, et al. Mitochondrial division occurs concurrently with autophagosome formation but independently of Drp1 during mitophagy. J Cell Biol. 2016;215(5):649–665.
- Oka S, et al. PPARα-Sirt1 complex mediates cardiac hypertrophy and failure through suppression of the ERR transcriptional pathway. Cell Metab. 2011;14(5):598–611.
-
Version history
- Version 1 (December 4, 2018): In-Press Preview
- Version 2 (January 22, 2019): Electronic publication
- Version 3 (February 1, 2019): Print issue publication



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)